Subcellular Targeted Metabolomics Service
Subcellular targeted metabolomics is a technology that utilizes high-throughput mass spectrometry to perform quantitative and qualitative analysis of metabolites within distinct subcellular structures (such as mitochondria, cytosol, nucleus, etc.). This service is based on high-sensitivity mass spectrometric analysis, combined with targeted metabolomics approaches, to ensure precise metabolite detection. It enables in-depth investigation of metabolic activities within specific intracellular regions and reveals the roles of various subcellular structures in cellular function and metabolic regulation, thereby providing detailed molecular-level data for research.
Subcellular targeted metabolomics service is widely applied in fields such as cancer research, neuroscience, metabolic diseases, and drug development. It offers deep insights into region-specific metabolic activities within cells, helping scientists to understand changes in metabolic networks and mechanisms of cellular regulation, thus supporting disease mechanism research and the development of precision therapeutic strategies.
Services at MtoZ Biolabs
Relying on advanced technology platforms, MtoZ Biolabs has launched the subcellular targeted metabolomics service focusing on analyzing the distribution and abundance changes of metabolites within distinct subcellular structures (such as mitochondria, nucleus, and endoplasmic reticulum). This service integrates efficient subcellular fractionation and enrichment workflows with precise targeted metabolite detection to generate region-specific metabolomic data, aiding in the elucidation of intracellular metabolic pathways and regulatory mechanisms. Commonly used technology platforms include, but are not limited to, the following:
Analysis Workflow
1. Sample Preparation and Cell Lysis
Cells are treated with mild lysis buffers to preserve the integrity of subcellular structures and prevent metabolite degradation.
2. Subcellular Fractionation
Specific subcellular compartments such as mitochondria, nucleus, and endoplasmic reticulum are efficiently isolated using differential centrifugation, density gradient centrifugation, or immunomagnetic bead-based methods.
3. Metabolite Extraction
Targeted subcellular structures are subjected to metabolite extraction using pre-chilled organic solvents (such as methanol or acetonitrile), rapidly quenching metabolic reactions and preserving the native metabolic state.
4. High-Resolution Mass Spectrometry Analysis
Appropriate platforms are selected based on metabolite characteristics to perform targeted analysis, achieving accurate qualitative and quantitative results.
5. Data Processing and Functional Interpretation
Standard databases and bioinformatics tools are employed to conduct pathway annotation, differential analysis, and subcellular localization-based functional interpretation.
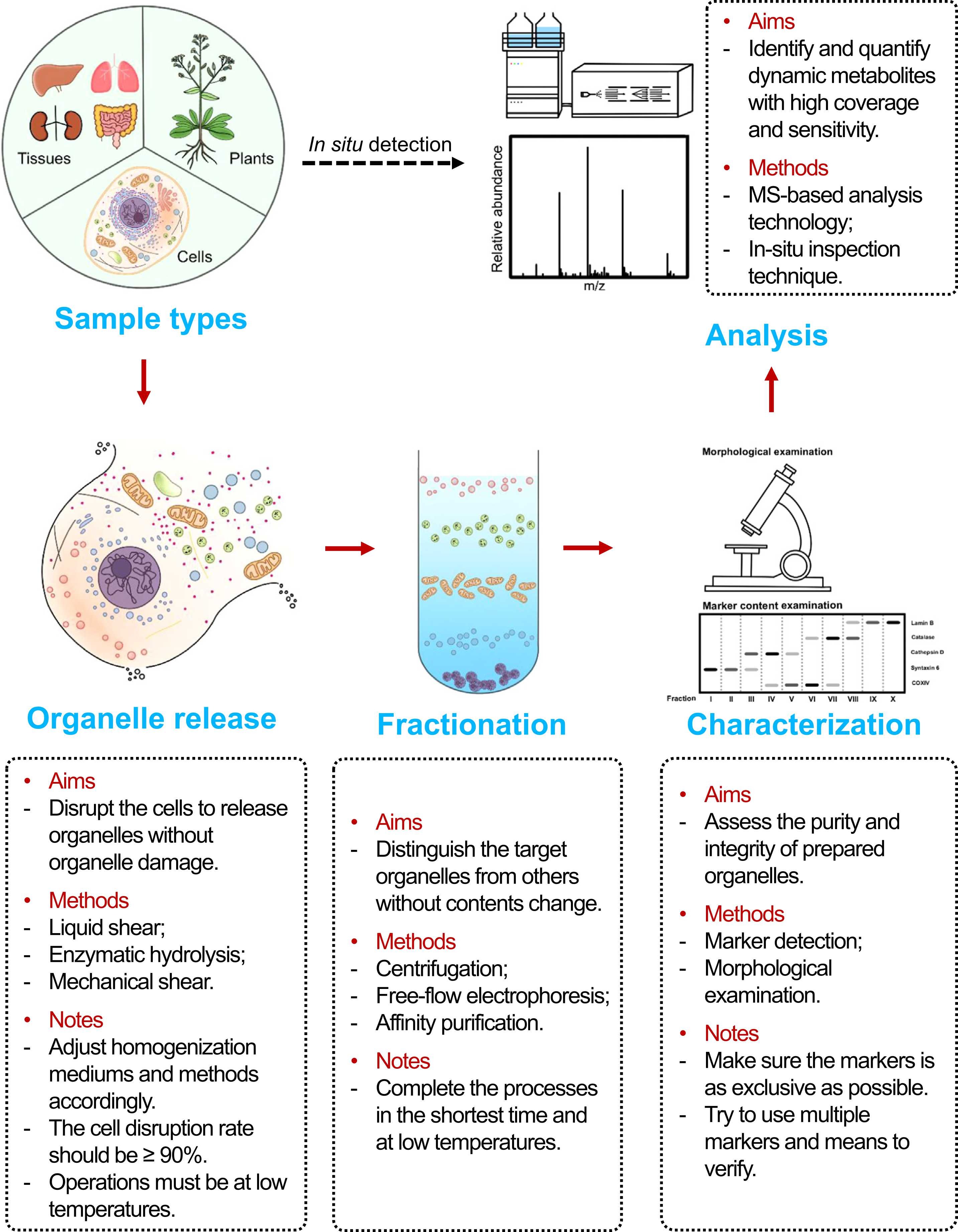
Qin, S Y. et al. Journal of Pharmaceutical and Biomedical Analysis, 2022.
Figure 1. Workflow for Subcellular Metabolomics.
Sample Submission Suggestions
1. Sample Types
Compatible with a wide range of samples including plant or animal tissues, cells, whole organisms, and biofluids. It is recommended to process all samples under sterile conditions and avoid repeated freeze–thaw cycles.
2. Sample Transportation
All samples must be shipped on dry ice to maintain low-temperature conditions throughout transit. For pre-extracted metabolites, please indicate the type and concentration of the extraction solvent used.
3. Additional Notes
If subcellular fractionation cannot be performed by the user, MtoZ Biolabs offers professional fractionation services. We also provide detailed sample submission guidelines and pre-processing recommendations—please contact us in advance for support.
Service Advantages
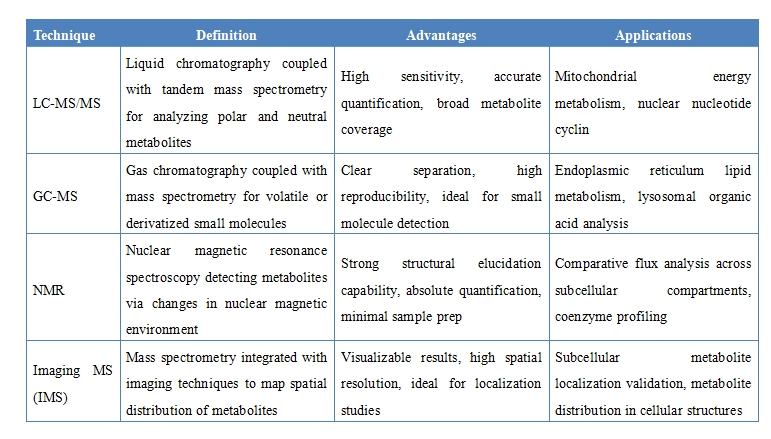
1. Multi-Platform Mass Spectrometry Support
Integrated use of advanced platforms including LC-MS/MS, GC-MS, NMR, and imaging mass spectrometry to accommodate metabolites with diverse physicochemical properties, offering high throughput, sensitivity, and qualitative capabilities.
2. High-Throughput Quantitative Analysis
Enables simultaneous detection of multiple targeted metabolites, suitable for complex samples and multi-component system-level studies, and ideal for large-scale experimental designs.
3. Expert Data Interpretation
Comprehensive bioinformatics support is provided, including pathway annotation, differential analysis, and functional enrichment, enhancing biological insight and data visualization.
4. Flexible Customization Options
Analytical strategies can be tailored to specific research goals and sample types, offering end-to-end scientific support from sample processing to data delivery, suitable for both basic research and translational medicine.
Applications
1. Cancer Metabolism Research
By analyzing metabolic reprogramming features within distinct subcellular compartments, this service helps identify metabolic vulnerabilities in cancer and supports the development of targeted therapeutic strategies.
2. Neurodegenerative Disease Mechanism Exploration
Subcellular targeted metabolomics service can be used to investigate energy metabolism disturbances in regions such as mitochondria and cytosol, aiding in the discovery of early metabolic biomarkers for diseases like Alzheimer’s and Parkinson’s.
3. Metabolic Disease Research
By examining subcellular metabolic pathway abnormalities in organs such as the liver and adipose tissue, this service contributes to understanding the pathogenesis of obesity, diabetes, and other metabolic disorders.
4. Cellular Function and Metabolic Regulation Studies
Subcellular targeted metabolomics service can be applied to study compartment-specific metabolic changes involved in biological processes such as cell proliferation, apoptosis, and autophagy, supporting in-depth research into molecular regulatory mechanisms.
FAQ
Q1: Which Subcellular Structures Can Be Analyzed for Metabolites?
A1: Commonly analyzable compartments include mitochondria, nucleus, plasma membrane, endoplasmic reticulum, peroxisomes, and Golgi apparatus. The detectability of specific structures depends on the sample type and the purity of the fractionation.
Q2: Will Subcellular Metabolite Extraction Alter the Metabolic State?
A2: We employ pre-chilled solvents and rapid quenching procedures to preserve the native metabolic state as much as possible. Additionally, strict quality control measures are implemented to minimize human-induced variability and ensure physiological relevance of the data.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Data Analysis, Preprocessing, and Estimation
4. Bioinformatics Analysis
5. Raw Data Files
How to order?







