Struggling with N-Terminal Protein Sequencing? 2 Common Issues and How to Fix Them!
In proteomics, N-terminal protein sequencing plays a critical role in elucidating the initiation sites of proteins, post-translational modifications (PTMs), signal peptide cleavage, and protein degradation pathways. However, a frequent technical challenge encountered by researchers is the inability to detect the N-terminal protein using standard methods.
This article explores the mechanistic basis of two major technical bottlenecks in N-terminal protein sequencing and presents practical solutions to enhance detection efficiency. These insights are particularly relevant for studies involving protein modification, degradation mechanisms, or antibody validation.
Issue 1: Chemical Blockage of the N-Terminal Protein Impairs Mass Spectrometry Identification
1. Underlying Mechanism: Post-Translational Modifications Obscure the N-Terminal Protein
In eukaryotic systems, newly synthesized proteins often undergo various post-translational modifications (PTMs) at the N-terminal protein, including:
(1) N-terminal acetylation
(2) Formylation
(3) Removal of the signal peptide or initial methionine (Met)
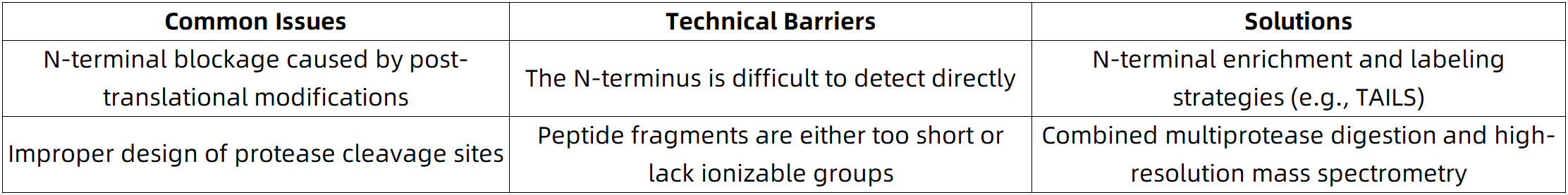
These modifications can neutralize the free amino group at the N-terminal protein, interfering with Edman degradation or mass spectrometry (MS)-based identification of the authentic N-terminal protein residue. This leads to reduced signal detection or incorrect assignment of the N-terminal protein.
2. Solution: Targeted Enrichment Strategies for the N-Terminal Protein
To improve detection sensitivity, current approaches utilize selective N-terminal protein labeling and enrichment techniques, such as:
(1) TAILS (Terminal Amine Isotopic Labeling of Substrates)
(2) COFRADIC (Combined Fractional Diagonal Chromatography)
(3) N-terminomics-specific reagents or tagging kits
These methods chemically block the internal peptide amines while enabling selective labeling and enrichment of genuine N-terminal protein peptides, thereby enhancing both signal strength and identification accuracy. At MtoZ Biolabs, we have developed an optimized platform that combines TAILS with high-resolution mass spectrometry, applicable to a wide range of biological samples including mammalian, yeast, and plant systems. This platform enables efficient characterization of N-terminal protein structures and their associated modifications.
Issue 2: Protease Cleavage Sites Located Near the N-Terminal Protein Can Lead to Loss of N-Terminal Peptides
1. Underlying Cause: Suboptimal Digestion Strategy Design
In most proteomics workflows, trypsin is the protease of choice due to its high specificity for lysine (K) and arginine (R) residues. However, the N-terminal region of proteins often lacks favorable cleavage sites for trypsin, which can result in:
(1) N-terminal peptides that are too short (<6 amino acids) to be effectively detected by mass spectrometry
(2) N-terminal peptides that lack sufficient charged residues, leading to poor ionization efficiency during MS analysis
This constitutes one of the key reasons why N-terminal protein signals are frequently undetectable in proteomics datasets.
2. Solution: Combinatorial Protease Digestion Strategy
To enhance the detection sensitivity for N-terminal protein peptides, the following strategies may be implemented:
(1) Utilization of alternative proteases with specific cleavage preferences, such as Lys-C, Asp-N, or Glu-C
(2) Application of sequential or combined enzymatic digestion protocols (e.g., Lys-C followed by trypsin)
(3) Optimization of digestion buffer pH to improve accessibility of cleavage sites
Moreover, coupling these strategies with high-sensitivity LC-MS/MS instrumentation and fine-tuned high-resolution MS acquisition parameters can further boost the detectability of N-terminal protein peptides. MtoZ Biolabs offers customizable multi-enzyme digestion workflows integrated with advanced platforms such as the Orbitrap Exploris 480, providing robust analytical support for applications including antibody drug characterization and identification of protein degradation targets.
Summary
Incomplete detection of the N-terminal protein remains a technical bottleneck but also presents an opportunity for methodological advancement.
With the increasing focus on post-translational modification (PTM) studies and the development of targeted protein degradation therapies, precise characterization of protein N-termini is becoming critically important. If you are initiating projects in this area, MtoZ Biolabs offers tailored N-terminal protein sequencing strategies, with comprehensive support covering the entire workflow—from sample preparation to data analysis. MtoZ Biolabs provides professional N-terminal protein sequencing services to a broad spectrum of clients. Our integrated workflow solution is designed to reduce experimental burden and improve research efficiency, enabling you to advance your N-terminal studies with greater speed and confidence.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







