Site-Specific Phosphorylation Modification Analysis Service
Site-Specific Phosphorylation Modification Analysis Service is designed for precise identification of protein phosphorylation sites. Phosphorylation primarily occurs on serine (Ser), threonine (Thr), and tyrosine (Tyr) residues, catalyzed by protein kinases and reversed by phosphatases, thereby modulating protein function. Site-specific phosphorylation has high specificity in regulating protein activity. Phosphorylation at different sites can lead to distinct conformational changes, interaction dynamics, or pathway activation. Therefore, accurately mapping phosphorylation sites and understanding their functional implications is essential.
Services at MtoZ Biolabs
1. Target Protein Site-Specific Phosphorylation Analysis
MtoZ Biolabs can perform site-level phosphorylation detection for specific target proteins, accurately identifying modification changes at single or multiple key sites under different conditions. Relying on a high-resolution LC-MS/MS platform, we provide researchers with clear information on site regulation to support functional mechanism validation.
2. Site-Specific Phosphoproteomics Analysis
MtoZ Biolabs applies phosphorylation enrichment strategies combined with mass spectrometry-based proteomics to systematically analyze site-specific modification patterns within large protein backgrounds. This analysis enables high-throughput site quantification and helps researchers comprehensively understand key phosphorylation events in signaling pathways and regulatory networks.
Analysis Workflow
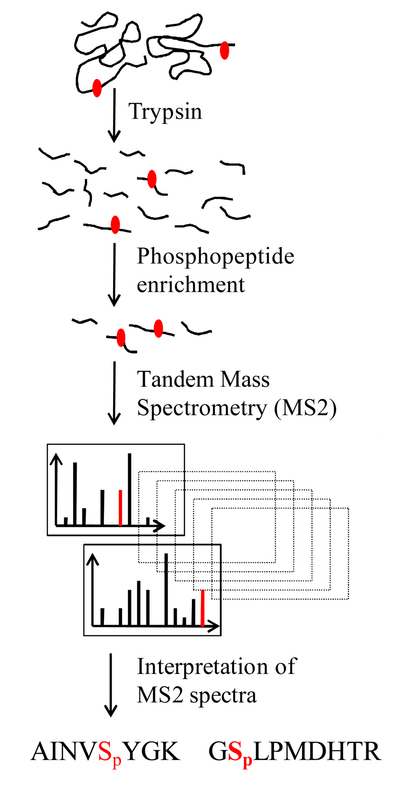
Mayya V. et al. Expert Review of Proteomics. 2009.
1. Sample Preparation
Protein samples are extracted, and phosphatase inhibitors are added to prevent the loss of phosphorylation modifications. Proteins are then enzymatically digested.
2. Phosphopeptide Enrichment
Selective enrichment of phosphorylated peptides is performed using IMAC (Fe³⁺, Ga³⁺), TiO₂, or a dual-enrichment strategy to maximize phosphopeptide capture.
3. High-Resolution Mass Spectrometry Analysis
Nano-LC is coupled with high-resolution mass spectrometry, employing HCD + ETD/EThcD fragmentation modes to enhance the accuracy of phosphorylation site identification.
4. Phosphorylation Site Identification
Phosphorylation sites are localized and validated using PhosphoRS, Ascore, and PTMProphet, ensuring high-confidence site assignment.
5. Bioinformatics Analysis
Comprehensive bioinformatics analysis includes pathway enrichment, kinase-substrate prediction, and protein-protein interaction (PPI) network construction, providing insights into the biological significance of phosphorylation modifications.
Applications
1. Cellular Signaling Pathway Research
Site-Specific Phosphorylation Modification Analysis enables the investigation of phosphorylation regulatory patterns in key signaling pathways such as PI3K-AKT, MAPK, mTOR, and JAK-STAT.
2. Disease Mechanism Research
Analyze aberrant phosphorylation signaling pathways in diseases such as diabetes and cancer, aiding in the identification of potential disease biomarkers.
3. Drug Target Screening
Site-Specific Phosphorylation Modification Analysis allows for monitoring the effects of kinase inhibitors on specific phosphorylation signals, supporting targeted drug discovery.
4. Immunology & Metabolic Regulation Research
Analyze the phosphorylation modifications of T cell receptor (TCR), B cell receptor (BCR), and AMPK metabolic signals to reveal the immune and metabolic regulation mechanisms.
FAQ
Q. How can the functional impact of site-specific phosphorylation be analyzed?
Understanding the role of a specific phosphorylation site requires bioinformatics analysis and experimental validation:
1. Use PhosphoSitePlus, NetPhos or GPS 5.0 to predict the potential upstream kinase of the site, and analyze its role in the signaling pathway in combination with the kinase family.
2. Perform KEGG/Reactome Pathway enrichment analysis to determine the biological significance of the phosphorylation modification in cell signaling pathways, metabolic regulation or disease progression.
3. Construct a protein interaction network (PPI) to analyze whether phosphorylation affects protein-protein interactions, and combine AlphaFold or PDB structure analysis to evaluate how the modification changes the protein conformation.
4. Perform experimental verification, such as disrupting upstream kinase expression through CRISPR-KO, or analyzing the direct effect of the phosphorylation site on protein function through point mutation (Ser/Thr/Tyr → Ala/Phe).
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs provides comprehensive phosphoproteomic services, including Site-Specific Phosphorylation Modification Analysis and phosphorylated protein quantitative analysis, and other analysis services related to phosphorylated proteins. Please feel free to contact us.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study utilized Site-Specific Phosphorylation Modification Analysis to identify site-specific phosphorylation modifications on key PSII subunits (e.g., D1, D2, CP43) and examined their dynamic changes following light-induced damage to PSII. The results revealed that site-specific phosphorylation of PSII subunits, such as D1-Ser331 and D2-Thr2, facilitates the disassembly of the protein complex, creating space for subsequent repair processes. Furthermore, the study found that dephosphorylation is a crucial step in PSII reassembly, ensuring that the restored photosystem can regain its photosynthetic activity.
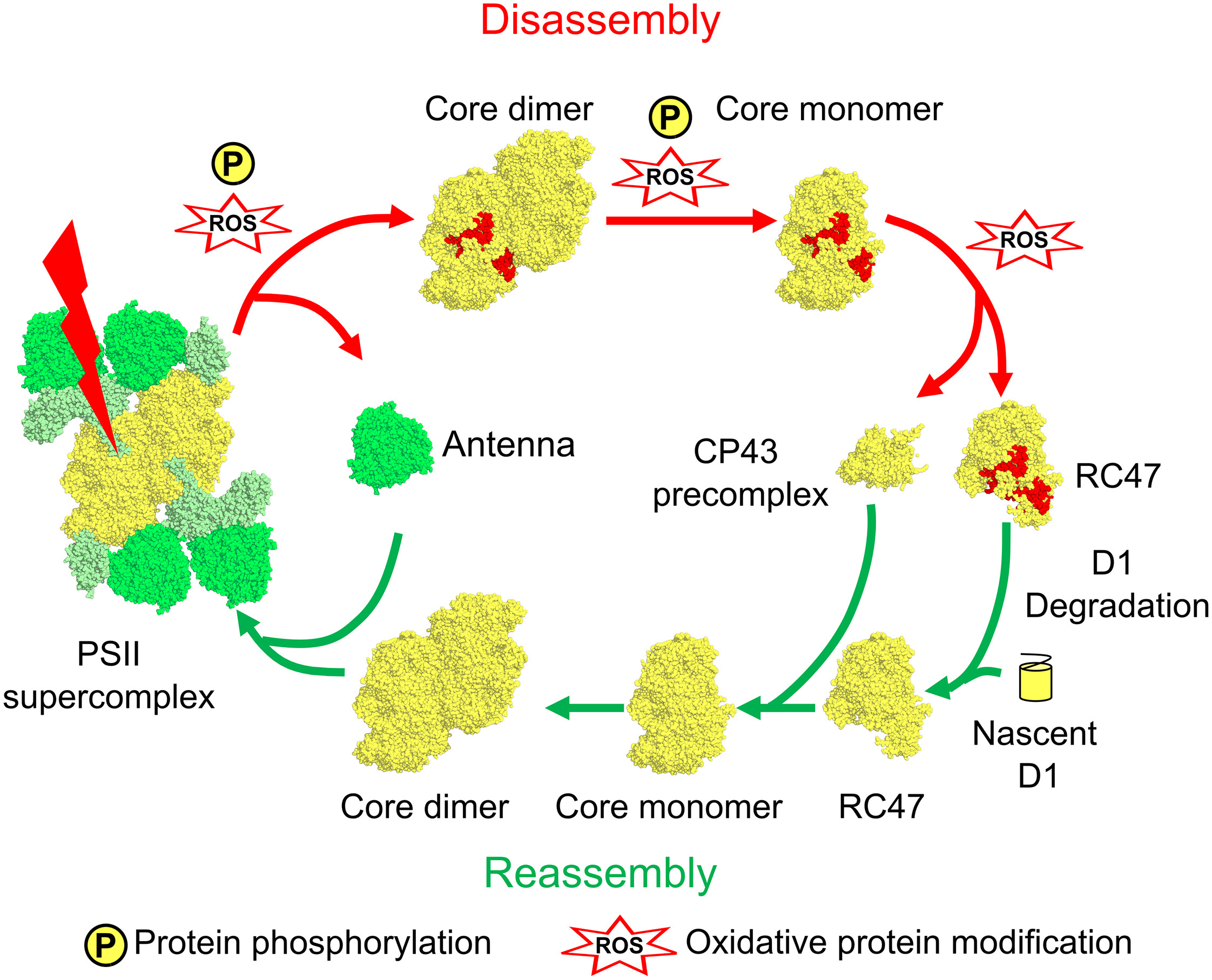
Mckenzie S D. et al. Plant Communications. 2024.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Quantitative Phosphoproteomics Service
How to order?







