Single-Cell & Tissue Exosome Combined Transcriptomic Research Service
Single-cell & tissue exosome combined transcriptomic research is an emerging approach that integrates single-cell transcriptomic sequencing with transcriptome profiling of tissue-derived exosomes. This strategy aims to elucidate the relationship between cellular states and exosome functions from multiple levels and dimensions. Utilizing platforms such as Genomics and high-throughput RNA sequencing technologies, this service captures the transcriptional landscapes of various cellular subpopulations within tissues, as well as RNA expression profiles in tissue exosomes. Through integrative analysis, it uncovers intercellular communication mechanisms, signaling pathways, and the functional roles of exosomes in disease initiation and progression.
Single-cell & tissue exosome combined transcriptomic research service is widely applied in tumor microenvironment studies, immune regulatory mechanism analysis, regenerative medicine, neurodegenerative disease research, and the investigation of stem cell therapy mechanisms. It is especially suitable for exploring the functional coupling between cellular behavior and exosome secretion in the context of complex tissue heterogeneity. This service provides critical data support for disease mechanism studies, biomarker discovery, and targeted therapy development.
Services at MtoZ Biolabs
Based on the integration of single-cell sequencing platforms and high-throughput RNA sequencing systems, and combined with techniques such as ultracentrifugation, the single-cell & tissue exosome combined transcriptomic research service provided by MtoZ Biolabs enables the isolation and extraction of both single cells and tissue-derived exosomes, yielding high-quality RNA data from both sources. The service includes tissue sample preprocessing, single-cell isolation and library construction, exosomal RNA extraction and library preparation, sequencing, and joint bioinformatics analysis. This ensures the acquisition of high-resolution transcriptional profiles of cellular subpopulations and exosomal transcriptome expression data, supporting mechanistic studies of the functional relationship between cellular states and exosome secretion.
Service Advantages
1. Multidimensional Integrated Analysis
Simultaneously obtains transcriptional profiles of diverse cellular subpopulations within tissues and RNA expression data from tissue-derived exosomes, enabling coordinated analysis of intracellular and extracellular gene expression. This approach reveals cell–cell communication mechanisms within complex microenvironments.
2. High Resolution and Sensitivity
Powered by single-cell sequencing platforms and high-throughput RNA sequencing systems, this service ensures data accuracy and depth, making it well-suited for low-input samples and the analysis of rare cell subpopulations.
3. Standardized End-to-End Workflow
Covers the entire process including tissue processing, single-cell isolation, exosome extraction, RNA library preparation, and integrated data analysis. This ensures high data comparability across samples and reproducibility of analytical results.
4. Customizable Research Design
Allows flexible selection of analytical content and sequencing depth based on project needs. Supports a wide range of research objectives such as disease mechanism investigation, biomarker discovery, and exosome functional studies.
Applications
1. Tumor Microenvironment Research
The single-cell & tissue exosome combined transcriptomic research service can be used to reveal the interactions between different cellular subpopulations in tumor tissues and their secreted exosomes, exploring key mechanisms of tumor initiation, metastasis, and immune evasion.
2. Immune Regulation Mechanism Analysis
By jointly analyzing the functional status of immune cells and the signaling mediated by their exosomes, this service elucidates their regulatory roles in processes such as inflammation and autoimmune diseases.
3. Stem Cell Therapy Mechanism Research
The single-cell & tissue exosome combined transcriptomic research service can be used to assess the distribution of stem cell subpopulations and the regulatory mechanisms of their secreted exosomes in tissue repair and regeneration, providing a theoretical basis for regenerative medicine.
4. Disease Biomarker Discovery
By jointly analyzing cell types with specific expression profiles and the RNA characteristics of their exosomes, this service supports the development of novel diagnostic or prognostic biomarkers.
Case Study
1. Unveiling the Role of PPIF and Macrophage Subtypes in LSCC Progression Via Single-Cell and Exosome RNA Sequencing
This study aims to investigate the role of mitochondrial protein PPIF and different macrophage subtypes in the progression of laryngeal squamous cell carcinoma (LSCC) through a combined analysis of single-cell RNA sequencing and tissue-derived exosome RNA sequencing. The study subjects included tumor tissues and corresponding exosome samples from LSCC patients. Single-cell RNA sequencing was used to analyze the heterogeneity of immune cells within the tumor microenvironment, while exosome RNA sequencing was employed to identify key genes present in tumor-derived exosomes. The results showed that PPIF was significantly overexpressed in M2-type macrophages and enriched in exosomes, suggesting that it may promote tumor immune evasion and metabolic reprogramming by regulating mitochondrial function and intercellular communication. The study concludes that integrated transcriptomic analysis of single cells and exosomes can systematically reveal tumor immune microenvironment dynamics and exosome-mediated regulatory mechanisms, highlighting PPIF as a potential diagnostic and therapeutic target for LSCC.
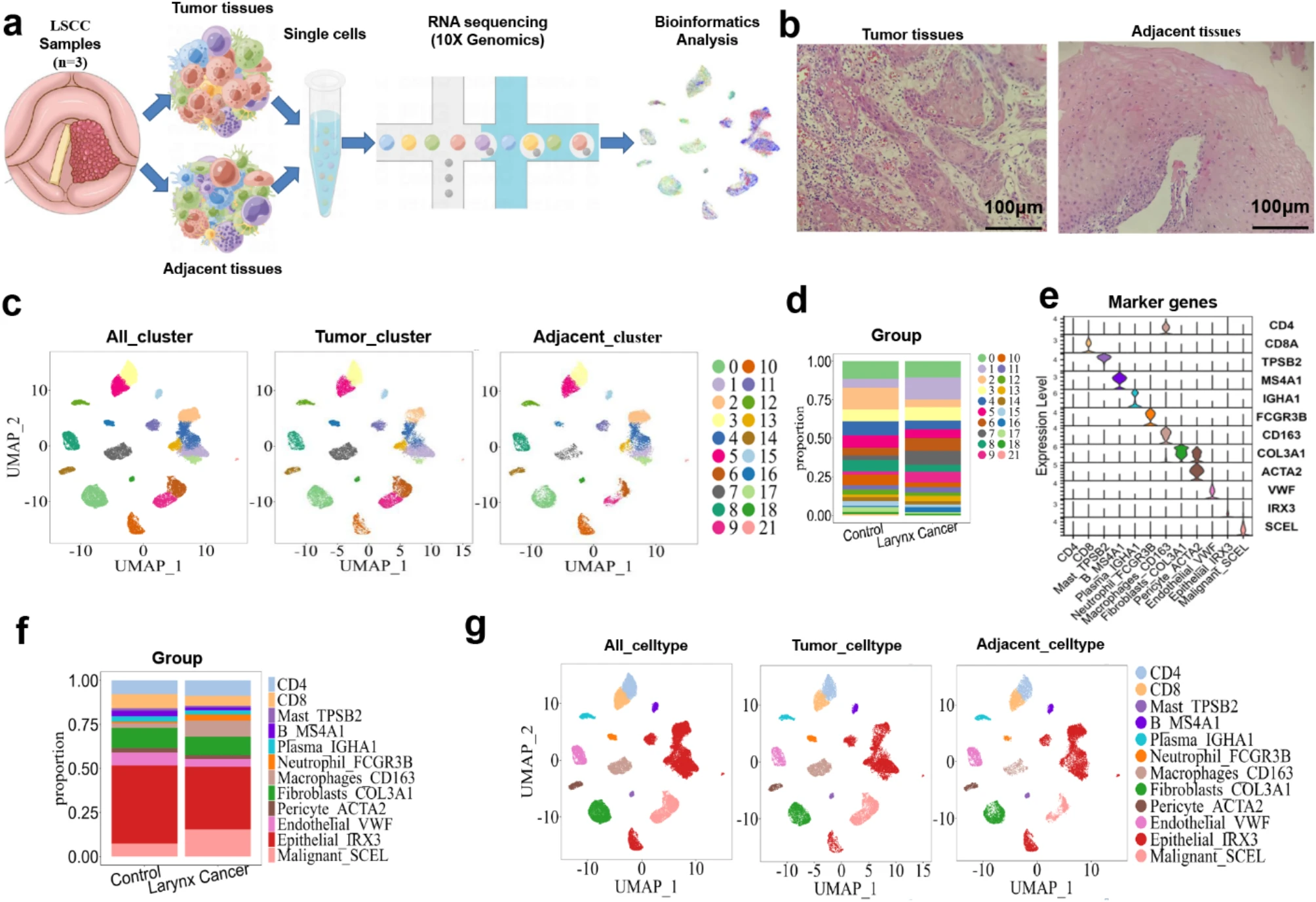
Wang, C J. et al. Scientific Reports, 2025.
Figure 1. Landscape View of Cell Composition in Tumor Tissue in Patients with LSCC.
FAQ
Q1: Can Single-Cell Sequencing and Exosome Analysis Be Performed Simultaneously?
A1: Yes. During the experimental design phase, we coordinate the tissue separation workflow to ensure synchronized extraction of single-cell and exosome samples. Subsequent bioinformatics analysis includes integrated data processing and association modeling.
Q2: Do you Support Customized Research Designs?
A2: Absolutely. We offer flexible customization of analysis content, sequencing depth, library preparation methods, and downstream analysis based on the research objectives, ensuring that the data output aligns with specific scientific needs.
How to order?







