Single-Cell & Exosome Combined Research Service
Single-cell & exosome combined research is a novel research strategy that integrates single-cell omics with exosome analysis, aiming to elucidate the relationship between cellular heterogeneity and exosome function at the individual cell level. This service utilizes technologies such as single-cell RNA sequencing (scRNA-seq) and single-cell secretion profiling, combined with exosome isolation, characterization, and molecular omics analysis, to achieve high-resolution, multidimensional studies of cell states, secretory behavior, and exosome cargo.
Single-cell & exosome combined research service is widely applied in tumor microenvironment research, immune regulation mechanism exploration, disease progression tracking, and stem cell therapy mechanism analysis. It is particularly suited for studying the synergistic roles of cells and exosomes in disease pathogenesis, signal transduction, and drug response. By integrating single-cell and exosome analyses, this service reveals the dynamic connections between cellular function and secreted vesicles, providing powerful technical support for precision medicine and mechanistic studies.
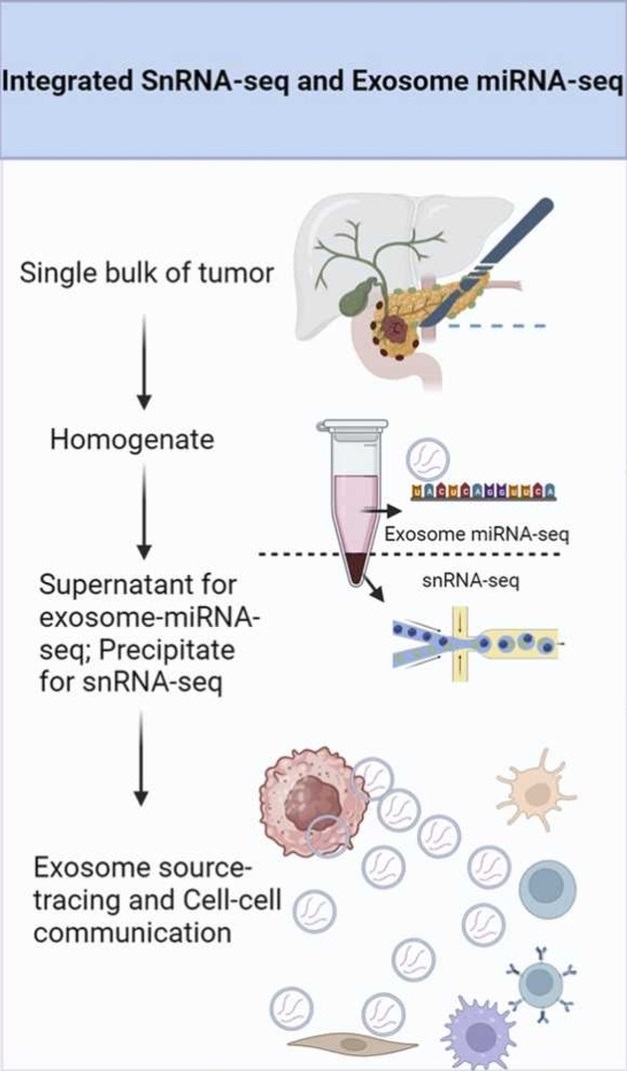
Tang, R. et al. Computational and Structural Biotechnology Journal, 2024.
Figure 1. Integrative Analysis of Single Cells and Exosomes.
Services at MtoZ Biolabs
Based on a single-cell sequencing platform combined with exosome isolation, purification, and multi-omics analysis technologies, the single-cell & exosome combined research service provided by MtoZ Biolabs enables coordinated investigation of individual cell states and their secreted exosomes. The service delivers high-resolution single-cell transcriptomic data along with detailed information on exosome particle size, concentration, and molecular characteristics, revealing the relationships between cellular functions and secreted vesicles while ensuring data accuracy, reproducibility, and interpretability.
Service Advantages
1. Multidimensional Integrated Analysis
Simultaneously obtain single-cell transcriptomic data and exosomal molecular features to comprehensively analyze the dynamic relationship between cellular functions and secretory behavior.
2. High Resolution and Sensitivity
Powered by the 10x Genomics platform and advanced instruments such as NTA and TEM, this service ensures precise and reproducible data, making it suitable for low-input samples.
3. Adaptable to Complex Systems
Ideal for studying highly heterogeneous tissues or microenvironments such as tumors, the immune system, and stem cells, enabling the discovery of key regulatory mechanisms.
4. Customizable Solutions
Offers flexible experimental design and data analysis strategies to meet diverse research goals for functional validation and mechanistic exploration.
Applications
1. Tumor Microenvironment Research
The single-cell & exosome combined research service can reveal the states of various tumor or immune cells and the roles of their secreted exosomes in promoting tumor progression and immune evasion.
2. Immune Regulation Mechanism Exploration
By analyzing immune cells, this service investigates their functional heterogeneity and the regulatory roles of their exosome secretion in inflammatory responses and autoimmune diseases.
3. Stem Cell Function and Secretome Analysis
The single-cell & exosome combined research service enables characterization of stem cell subpopulations and exploration of the mechanisms by which their exosomes contribute to tissue repair and regeneration.
4. Disease Progression and Biomarker Discovery
By integrating single-cell and exosome data from pathological tissues, this service helps identify early disease biomarkers and potential therapeutic targets.
Case Study
1. Combining Single-Cell RNA Sequencing of Peripheral Blood Mononuclear Cells and Exosomal Transcriptome to Reveal the Cellular and Genetic Profiles in COPD
This study aims to reveal the cellular and genetic characteristics of patients with chronic obstructive pulmonary disease (COPD) by integrating single-cell RNA sequencing of peripheral blood mononuclear cells (PBMCs) and exosomal transcriptome analysis. The research subjects include PBMC samples and plasma-derived exosomes from COPD patients and healthy controls. Methodologically, single-cell RNA sequencing was performed on PBMCs to analyze cell subpopulation composition and gene expression profiles; meanwhile, transcriptome sequencing of plasma exosomes was conducted to examine their RNA content. The results showed that specific changes in the proportions of cell subpopulations and gene expression differences were observed in the PBMCs of COPD patients; exosomal transcriptome analysis revealed abnormally expressed RNA molecules related to inflammation and immune regulation in COPD patients. The study concludes that the combined analysis of single-cell RNA sequencing and exosomal transcriptomics can provide in-depth insights into the cellular and molecular features of COPD, offering a new perspective for understanding its pathogenesis and identifying potential biomarkers.
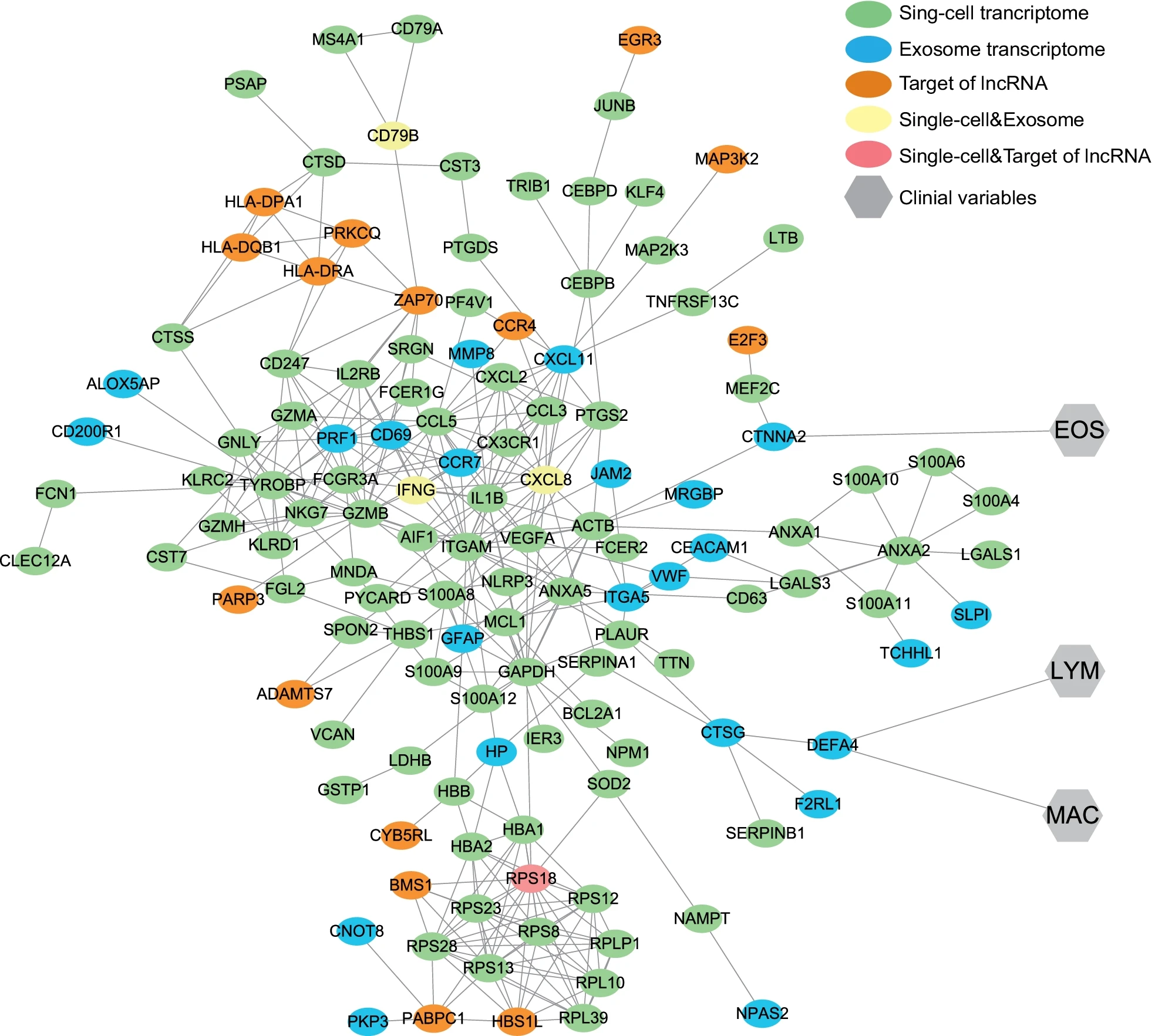
Pei, Y L. et al. Respiratory Research, 2022.
Figure 2. Protein–Protein Interaction Network and its Connection with Sputum Clinical Variables.
FAQ
Q1: Can Single-Cell Sequencing and Exosome Analysis Be Performed Simultaneously?
A1: Yes. During the sample preprocessing stage, we carry out single-cell isolation and exosome extraction in parallel to ensure high comparability between the two data systems, which facilitates downstream functional correlation analysis.
Q2: How Is the Data Integrated and Analyzed?
A2: We integrate single-cell transcriptomic data with exosome multi-omics data through bioinformatic analysis, applying feature correlation analysis and cell–vesicle relationship modeling to uncover functional links between cellular states and exosome secretion.
How to order?







