PTM Qualitative Analysis Service
PTM Qualitative Analysis Service utilizes high-resolution mass spectrometry to precisely identify post-translational modifications (PTMs) of proteins, providing information on modification types and modification sites. PTM of proteins is an important part of protein function regulation. It affects the stability, activity, interaction and subcellular localization of proteins by adding various chemical modifications to the amino acid residues of proteins. PTMs play a crucial role in cellular signal transduction, gene regulation, metabolic control, immune response, and other biological processes. Accurate identification and qualitative analysis of PTMs are essential for understanding cellular regulatory mechanisms, discovering biomarkers, and advancing precision medicine.
Leveraging advanced chromatography and mass spectrometry platforms, MtoZ Biolabs offers PTM Qualitative Analysis Service covering a broad range of modification types, including phosphorylation, acetylation, methylation, glycosylation, ubiquitination, and disulfide bond formation. Our service provides accurate and reliable protein modification identification solutions, ensuring high-quality data to support your research needs.
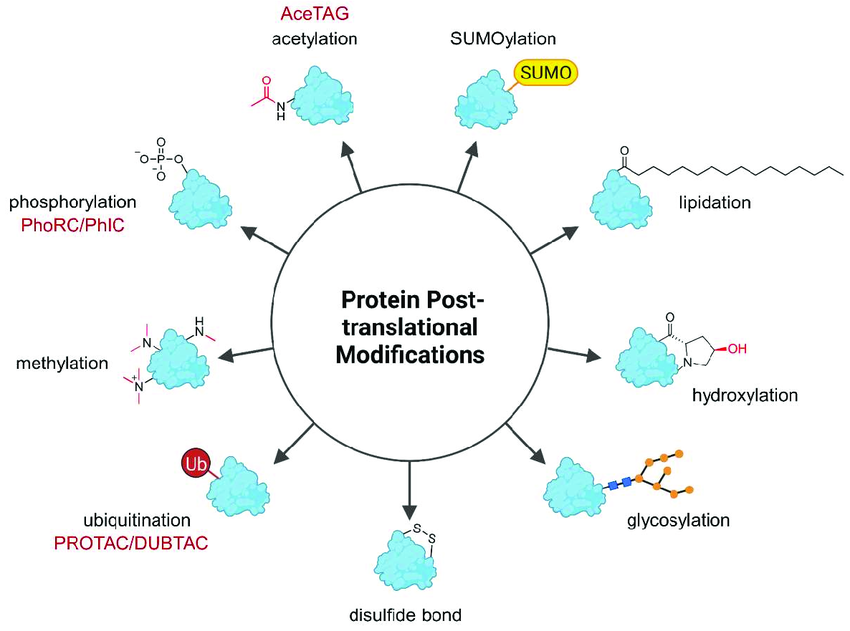
Heitel P. Molecules. 2023.
Analysis Workflow
1. Sample Preparation
Proteins are extracted and quantified from the sample, followed by digestion using trypsin or other specific proteases.
2. Modified Peptide Enrichment
Specific enrichment method is selected based on the target modification type. For example, phosphorylation peptides can be enriched using IMAC/TiO₂ affinity, acetylated/methylated peptides can be enriched using antibody affinity, and glycosylated peptides can be enriched using lectin affinity/HILIC separation, among others.
3. LC-MS/MS Analysis
Separation and detection are performed using ultra-high-performance liquid chromatography (UHPLC) and high-resolution mass spectrometry (Orbitrap/Q-TOF).
4. Data Analysis
Modified peptides are identified and bioinformatics analysis is conducted using databases and software tools.
Applications
Cell Signaling Pathway Research
Analyze the role of protein modifications in signal transduction, such as phosphorylation regulation in the MAPK and PI3K/Akt pathways.
Disease Biomarker Screening
Identify disease-associated protein modification patterns through PTM qualitative analysis.
Drug Target Research and Protein Drug Quality Control
Evaluate the impact of drugs on protein modification states, such as the regulation of kinase substrate phosphorylation by small-molecule inhibitors.
Analyze the PTM status of protein drug to ensure the expected biological activity and structural integrity.
Immunity and Inflammatory Response Mechanism Research
Investigate the regulatory roles of modifications such as ubiquitination and acetylation in immune responses.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced PTM Qualitative Analysis Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all PTM Qualitative Analysis Service data, providing clients with a comprehensive data report.
4. Comprehensive PTM Analysis: Covers a wide range of modification types, including phosphorylation, acetylation, glycosylation, ubiquitination, lipidation, and methylation, to meet diverse research needs.
FAQ
Q. How can protein sample extraction be optimized to maximize PTM preservation?
PTMs are highly sensitive to environmental factors such as temperature, pH, and enzymatic activity during protein extraction and preparation, which may lead to modification loss or non-specific modifications. Therefore, optimizing the protein extraction process is crucial:
1. Use Appropriate Buffers: Avoid high-salt and highly alkaline environments. It is recommended to use phosphate-buffered saline (PBS) or HEPES buffer combined with protease and phosphatase inhibitors (e.g., NaF, β-glycerophosphate, protease inhibitor cocktails) to prevent modification degradation.
2. Low-Temperature Processing: All experimental steps, including cell lysis, centrifugation, and ultrafiltration, should be performed at 4°C to minimize protein degradation.
3. Non-Denaturing Lysis Strategies: For modifications such as phosphorylation and glycosylation, avoid high concentrations of denaturants like SDS or urea to maintain the native protein-modification state.
4. Immediate Fixation: For unstable PTMs, such as ubiquitination related to proteasomal degradation, stabilization methods like cross-linking (e.g., formaldehyde, glutaraldehyde) or rapid freezing (liquid nitrogen snap-freezing) can be employed to preserve modification states.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study utilized high-resolution ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) to analyze PTM patterns in breast cancer and ovarian cancer patients. A total of 12 unique modified peptide segments from 8 proteins were identified in ovarian cancer patients, while 6 unique modified peptide segments from 3 proteins were detected in breast cancer patients. By integrating molecular dynamics simulations, researchers discovered that cancer-specific PTM may induce conformational changes in protein structures, thereby regulating their molecular functions and altering metabolic pathway characteristics.
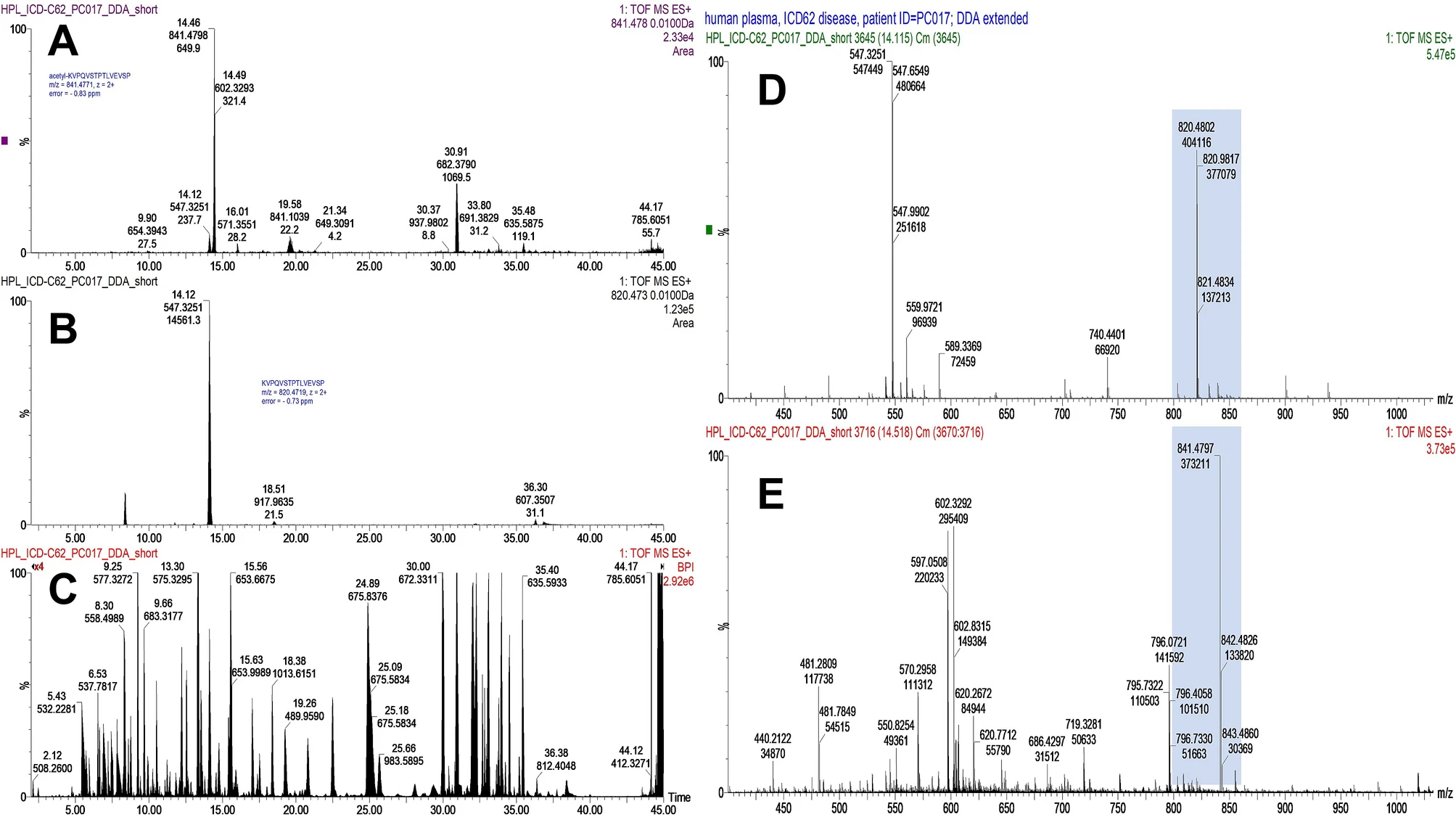
Tikhonov D. et al. Sci Rep. 2021.
How to order?







