Protein Subcellular Localization Service
Protein subcellular localization is critical for deciphering biological function. It describes the spatial distribution of a protein across cellular compartments, including nucleus, mitochondria, plasma membrane, endoplasmic reticulum, Golgi apparatus, lysosome, cytosol, and more. A protein’s activity, signaling role, interactions, and stability all depend on its precise location. Mis‑localization has been linked to cancers, neurodegenerative disorders, metabolic syndromes, and other diseases. Comprehensive localization profiling therefore advances basic biology and provides invaluable insights for target discovery and precision therapy.
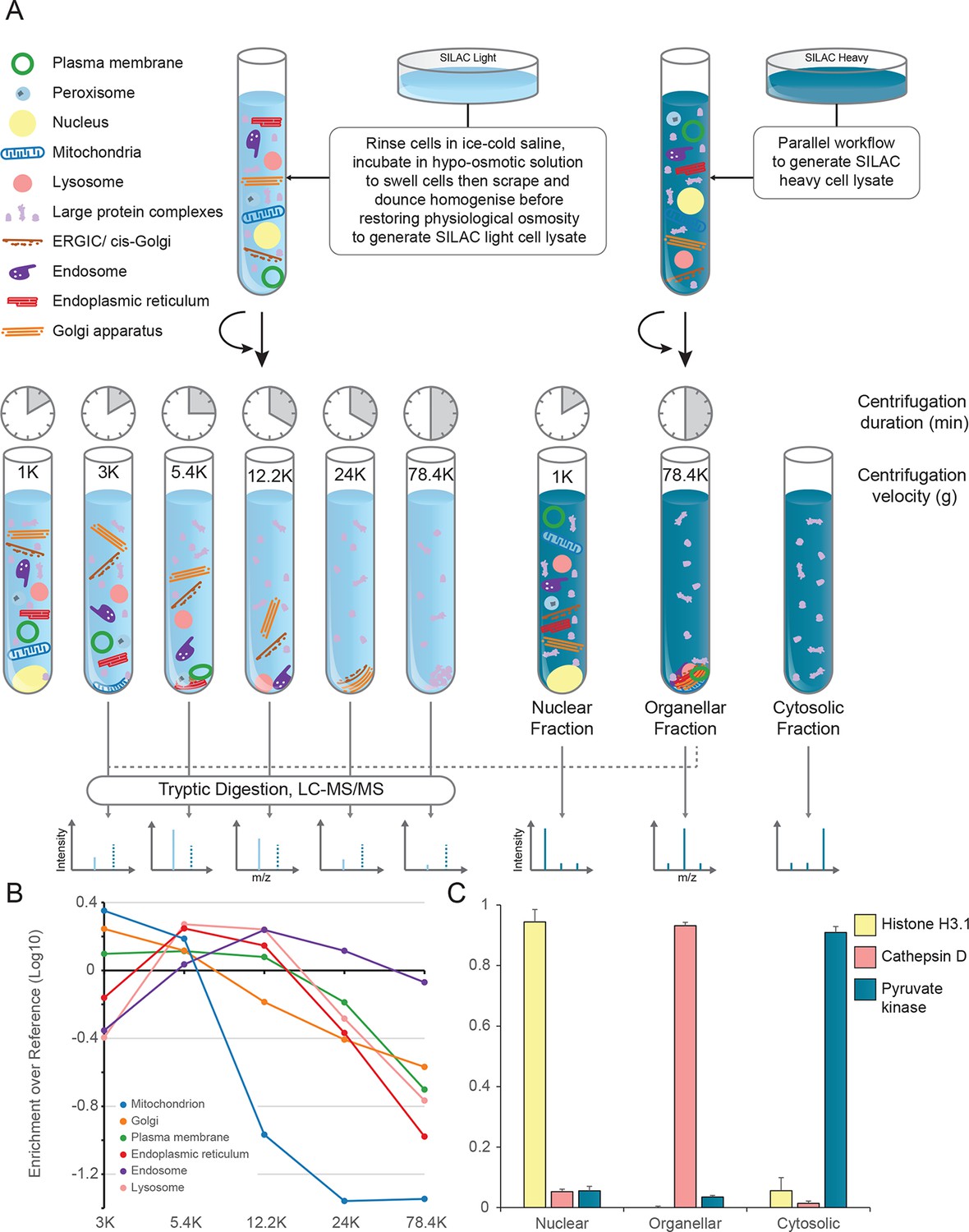
Itzhak, DN. et al. Elife. 2016.
Figure 1. Generation of Organellar Maps through Fractionation Profiling
Leveraging advanced fractionation technologies, high‑resolution mass‑spectrometry (MS) platforms, and powerful bioinformatics, MtoZ Biolabs has launched the Protein Subcellular Localization Service. Designed for accuracy, high throughput, and systematic coverage, this service delivers a panoramic view of protein localization, accelerating biomedical research and drug development.
Services at MtoZ Biolabs
Built on high‑sensitivity LC‑MS/MS, classical subcellular fractionation, and state‑of‑the‑art quantitative strategies (TMT, SILAC, DIA), the service enables quantitative detection and spatial inference of proteins across multiple compartments. Differential centrifugation, density gradients, and affinity‑bead sorting efficiently isolate nuclei, cytosol, mitochondria, ER, lysosomes, and more, with marker‑protein validation to confirm purity. Key Protein Subcellular Localization Service includes:
1. Subcellular Fractionation and Purity Validation
Optimal physical or immuno‑affinity methods selected per sample type for compartment‑specific isolation.
2. Quantitative Proteomics
Orbitrap Fusion Lumos, timsTOF HT, and nano‑LC systems combined with TMT or DIA deliver deep, precise coverage.
3. Localization Modeling and Annotation
Statistical scoring of spatial distributions, cross‑validated with public databases.
4. Dynamic Localization Tracking
Drug‑stimulus or time‑course designs reveal protein translocation and regulatory mechanisms.
5. Data Interpretation and Visualization
Enrichment analysis, network diagrams, chord plots, heat‑maps, and more for intuitive results display.
Analysis Workflow
1. Sample preparation and QC
2. Subcellular fractionation (differential centrifugation / gradient / immuno‑affinity)
3. Protein extraction, quant‑labeling, enzymatic digestion
4. LC‑MS/MS acquisition
5. Data processing, spatial prediction & annotation
6. Visualization & report delivery
Service Advantages
1. High‑Resolution Separation
Our protein subcellular localization service integrates centrifugation, gradients, and marker‑antibody magnetic beads, ensuring purity and minimize cross‑contamination.
2. Premium Analytical Platform
Thermo Orbitrap Fusion Lumos and Bruker timsTOF HT with nano‑LC achieve sensitive detection of low‑abundance proteins, ideal for large‑scale localization studies.
3. Customizable Service
MtoZ Biolabs supports static localization or dynamic tracking under stimulation, drug treatment, or phenotypic variation, flexibly meeting diverse research needs.
Applications
1. Cancer Biology: Investigate mis‑localized signaling proteins driving tumor progression.
2. Neurodegeneration: Monitor localization shifts of mitochondrial or lysosomal enzymes.
3. Drug Mechanism of Action: Confirm drug‑induced protein relocalization and pathway modulation.
4. Signal Transduction: Track stimulus‑dependent translocation of transcription factors and kinases.
5. Protein Function Annotation: Provide spatial context for newly discovered proteins.
FAQs
Q1. What sample types are accepted?
Cultured cells, tissue specimens, primary cells, and FFPE sections; please discuss tissue samples in advance to optimize experimental design.
Q2. How is compartment purity verified?
Optional Western blot or MS‑based marker analysis evaluates purity of each fraction.
Q3. What is the typical turnaround time of Protein Subcellular Localization Service?
Standard projects require approximately 4–5 weeks, subject to sample type and analysis depth.
Sample Submission Suggestions
1. Cells: ≥ 1 × 10⁷ cells, ship on dry ice.
2. Tissues: ≥ 50 mg, avoid freeze–thaw cycles.
3. Buffers: Exclude SDS, glycerol, high salt; include protease inhibitors if possible.
4. Metadata: Complete sample information sheet specifying treatment conditions and study objectives.
Deliverables
1. Fraction purity and QC report (optional WB or MS marker scores)
2. Quantitative protein data with localization confidence scores
3. Differential localization statistics and enrichment results
4. Bioinformatics report with figures (heat‑maps, chord plots, networks, etc.)
5. Interactive localization datasets (compatible with Cytoscape, R, Python)
Subcellular localization is a pivotal gateway to understanding protein function and disease mechanisms. MtoZ Biolabs combines expert fractionation, multi‑omics integration, and intuitive data visualization to provide a robust Protein Subcellular Localization Service. Contact us today to advance the frontiers of precision biology together!
How to order?







