Protein Seq Service
Protein sequences are essentially strings of amino acids linked in a specific order, determined by the genetic information within an organism. This arrangement, known as the protein sequence, dictates the three-dimensional structure of the protein and defines its function. Protein synthesis begins with DNA transcription, during which fragments of DNA are transcribed into mRNA (messenger RNA) within the cell nucleus, and then translated into amino acid chains. Each set of three nucleotides in the mRNA, referred to as a codon, corresponds to a specific amino acid. For instance, the codon AUG encodes methionine. This genetic code is nearly universal across all organisms, underscoring the fundamental role of proteins in life. Understanding protein sequences is crucial, as variations in these sequences can alter protein structure and function, potentially leading to disease. For example, a single change in the hemoglobin protein sequence results in sickle cell anemia, demonstrating how sequence variation directly affects protein functionality.
Protein seq service primarily relies on two methods: Edman degradation (Harvey and Ferrier, 2011; Bauer et al., 1997) and mass spectrometry (Sahukar et al., 2016). Currently, mass spectrometry is the most widely used method for protein sequencing and identification, while Edman degradation remains a key technique for characterizing the N-terminus of proteins. The general process of Edman degradation-based protein seq service involves reacting the protein with phenyl isothiocyanate under mildly alkaline conditions. The reaction releases the N-terminal residue from the polypeptide chain as a phenylthiohydantoin derivative of the amino acid, which is then analyzed (typically via liquid chromatography). The remaining peptide chain undergoes subsequent cycles of the reaction to determine the complete sequence.
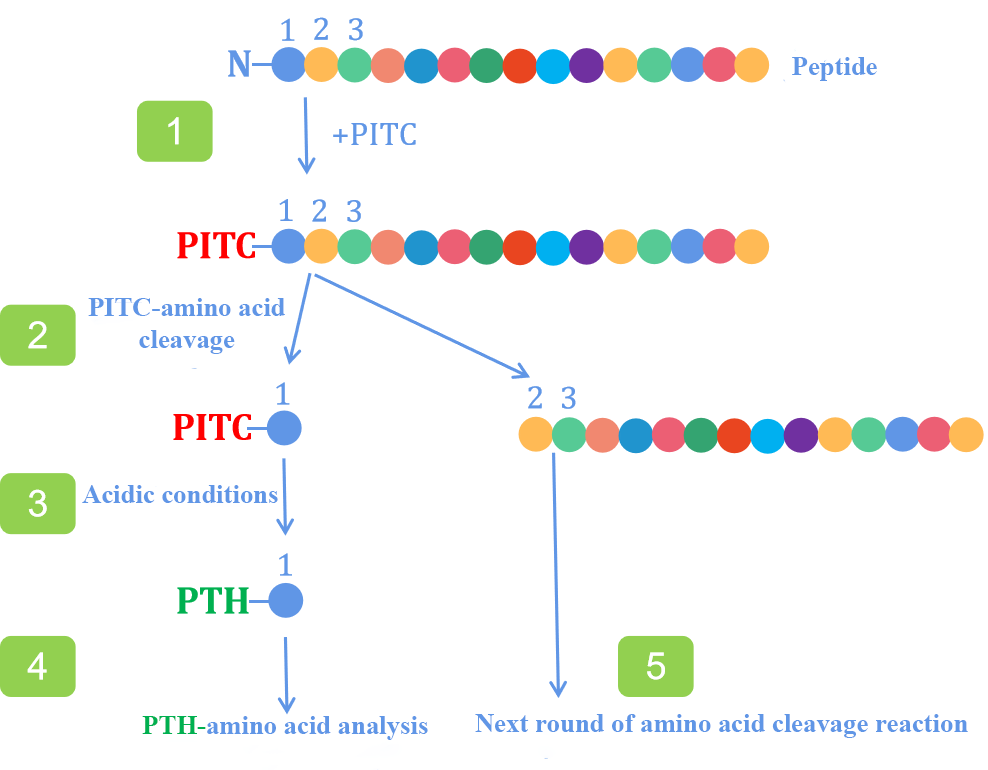
Figure 1. Edman Degradation Process.
Analyzing the N- and C-terminal sequences of proteins is an essential detection method in the pharmaceutical industry.At present, most N-terminal sequence analysis uses the Edman degradation method, but the Edman degradation method cannot solve the sequencing problems of N-terminal blocking and protein modification, while mass spectrometry sequencing is not limited. The C-terminal analysis of proteins cannot adopt chemical analysis similar to N-terminal amino acid sequence analysis. Currently, mass spectrometry is used to analyze the C-terminal sequence of protein samples. The development of matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) in the late 1980s brought a revolutionary leap in the application of mass spectrometry technology in protein structure analysis.Tandem mass spectrometry (MS/MS) is used to determine the molecular mass of peptide fragments obtained during mass spectrometry analysis. Peptide fragmentation in mass spectrometry follows specific patterns, with peptide bonds being the most prone to cleavage. This fragmentation generates ion species, typically referred to as b-ions and y-ions, which are then plotted on an m/z spectrum. By analyzing this spectrum, the N-terminal and C-terminal sequences of proteins can be deduced.
Services at MtoZ Biolabs
Based on nano LC-MS/MS combined with tandem mass spectrometry and Shimadzu Edman degradation sequencing system, the protein sequencing service platform established by MtoZ Biolabs can meet a variety of protein sequence analysis requirements. These include amino acid composition analysis, N-terminal sequencing, C-terminal sequencing, full-sequence analysis, and Edman degradation-based N-terminal sequencing. For proteins without theoretical sequences, MtoZ Biolabs offers de novo sequencing services to analyze unknown protein sequences.
Compared to Edman degradation, MS offers advantages such as higher sensitivity, rapid peptide fragmentation, and the ability to identify proteins with blocked or modified termini. Utilizing the existing high-resolution MS technology platform, the protein seq service provided by MtoZ Biolabs can achieve 100% coverage of the measured target protein sequence.These analyses are instrumental in confirming whether recombinant proteins are fully expressed and detecting any cleavage events during expression.
Service Advantages
1. Advanced Analytical Platform
MtoZ Biolabs has established advanced protein seq service platform, ensuring reliable, fast, and precise analyses.
2. Transparent Pricing
Our pricing is transparent, with no hidden or additional fees.
3. High-Quality Data
Our analyses feature deep data coverage, rigorous quality control, and comprehensive reporting.
4. Customized Research Solutions
MtoZ Biolabs can provide you with customized services to solve your unique research questions and experimental requirements.
Sample Submission Suggestions
MtoZ Biolabs provides one-stop protein seq service for samples in various forms, including PVDF membranes, gel spots, gel bands, and solution-phase samples.
Note: For specific requirements or assistance with sample preparation, please contact us.
Applications
1. Drug Design
By analyzing protein sequences, the interaction between drugs and target proteins can be predicted, aiding in the optimization of drug design.
2. Disease Prediction and Diagnosis
Protein seq service can help identify functional abnormalities and mutations in proteins, enabling disease risk prediction and diagnostic applications.
3. Evolutionary Study
Comparing protein sequences across species can elucidate evolutionary relationships, species histories, and gene family origins.
4. Protein Engineering
Insights into protein sequence and structure enable engineering modifications to enhance protein characteristics and functionality, with applications in biotechnology and biopharmaceuticals.
5. Novel Protein Discovery
The analysis of unknown biomolecular structure through protein seq service can discover new proteins, explore their structures and functions, and provide new ideas for the development of new drugs and biomaterials.
Case Study
1. Regulation of the Phage Defense System CBASS by a Prokaryotic E2 Enzyme Mimicking Ubiquitination
The cyclic oligonucleotide-based antiphage signaling system (CBASS) is an innate prokaryotic immune system composed of CBASS-associated proteins that leverage cyclic oligonucleotides to activate antiviral immunity. This study utilized biochemical, genetic, cryo-electron microscopy, and mass spectrometry analyses to demonstrate how the E2 enzyme in Serratia marcescens mimics a ubiquitination cascade to regulate cGAS. This includes processing of the cGAS C terminus, conjugation of cGAS to cysteine residues, linkage of cGAS to lysine residues, cleavage of isopeptide bonds, and polycGASylation. Polymerization of cGAS activates cGAS to produce cGAMP, which can serve as an antiviral signal and lead to cell death. These results uncover the unique regulatory role of E2 in the CBASS pathway.
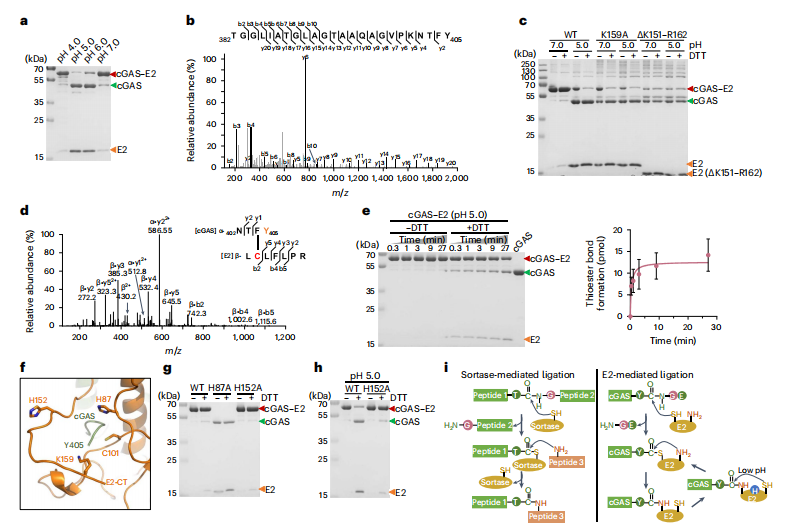
Yan, Y. et al. Nature microbiology, 2024.
Figure 2. The Role of E2 as A Cysteine Protease.
2. Bioengineered Human Serum Albumin Fusion Protein as a Targeted Tumor Therapeutic/Enzyme/pH Triple-Responsive Drug Delivery Vehicle
Human serum albumin (HSA) has the characteristics of good biocompatibility and long circulation time, and is widely used as a carrier for poorly soluble anticancer drugs. However, it also has shortcomings such as poor tumor targeting and uncontrollable drug release. This study uses gene fusion technology to introduce polyhistidine (pHis), matrix metalloproteinase 2 (MMP-2) digestion site, and arginine-glycine-aspartate (RGD) peptide at the separation end of HSA. HSA is modified to improve its biological properties. The resulting protein was expressed in Pichia pastoris, self-assembled into 3RGD-HSA-MMP-18His nanoparticles (RHMH18 NPs), and loaded with the hydrophobic drug paclitaxel (PTX) in the polyhistidine micelle core. RHMH18 NPs specifically bind to the ανβ3 integrin upregulated on tumor vascular endothelium through RGD, showing efficient active tumor targeting, resulting in the enrichment of therapeutic substances at the tumor site. After reaching the tumor microenvironment, RHMH18 NPs are cut off by MMP-2, removing the HSA-3RGD part, leaving small and positively charged histidine micelles, which can more effectively penetrate deep into the tumor tissue. Finally, the histidine micelles successfully escaped from the lysosomes and released the drug under the influence of pH. In vivo experimental results show that the three-stage propulsion RHMH18 NPs exhibit excellent tumor inhibitory activity with few side effects, providing a potential protein-based drug delivery system strategy for tumor treatment.
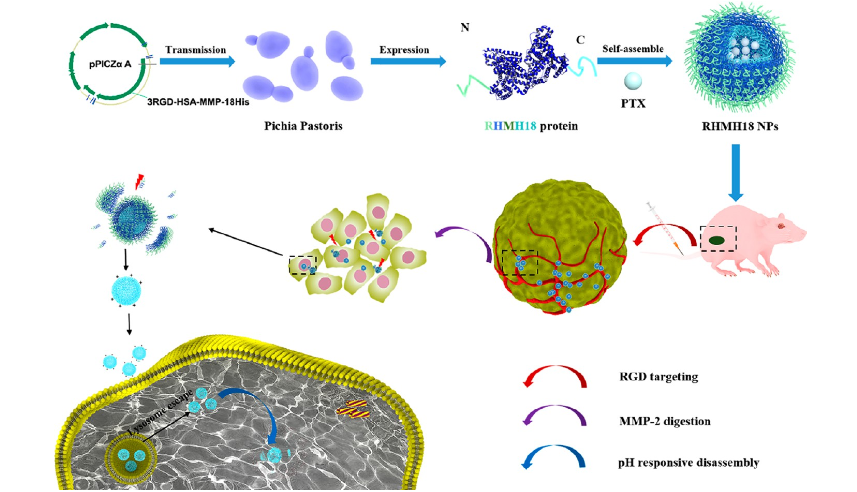
Wang, MY. et al. ACS Nano, 2020.
Figure 3. Schematic Route of HSA Fusion Protein NPs (RHMH18 NPs) and Its Application in Tumor Targeted Therapy.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Data Analysis, Preprocessing, and Estimation
4. Raw Data Files
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
De Novo Protein Sequencing Service
How to order?







