Protein PTM Crosstalk Analysis Service
Protein PTM Crosstalk Analysis Service is a mass spectrometry-based systematic service that focuses on exploring the interactions—synergy, competition, and dependence—between various post-translational modifications (PTMs) on proteins. Increasing evidence suggests that multiple PTMs can coexist or mutually exclude each other on the same protein or protein complex, forming regulatory networks involved in disease progression. This “crosstalk” phenomenon between PTMs not only greatly enhances the diversity of protein functions but also deepens our understanding of signaling pathways and cellular regulation.
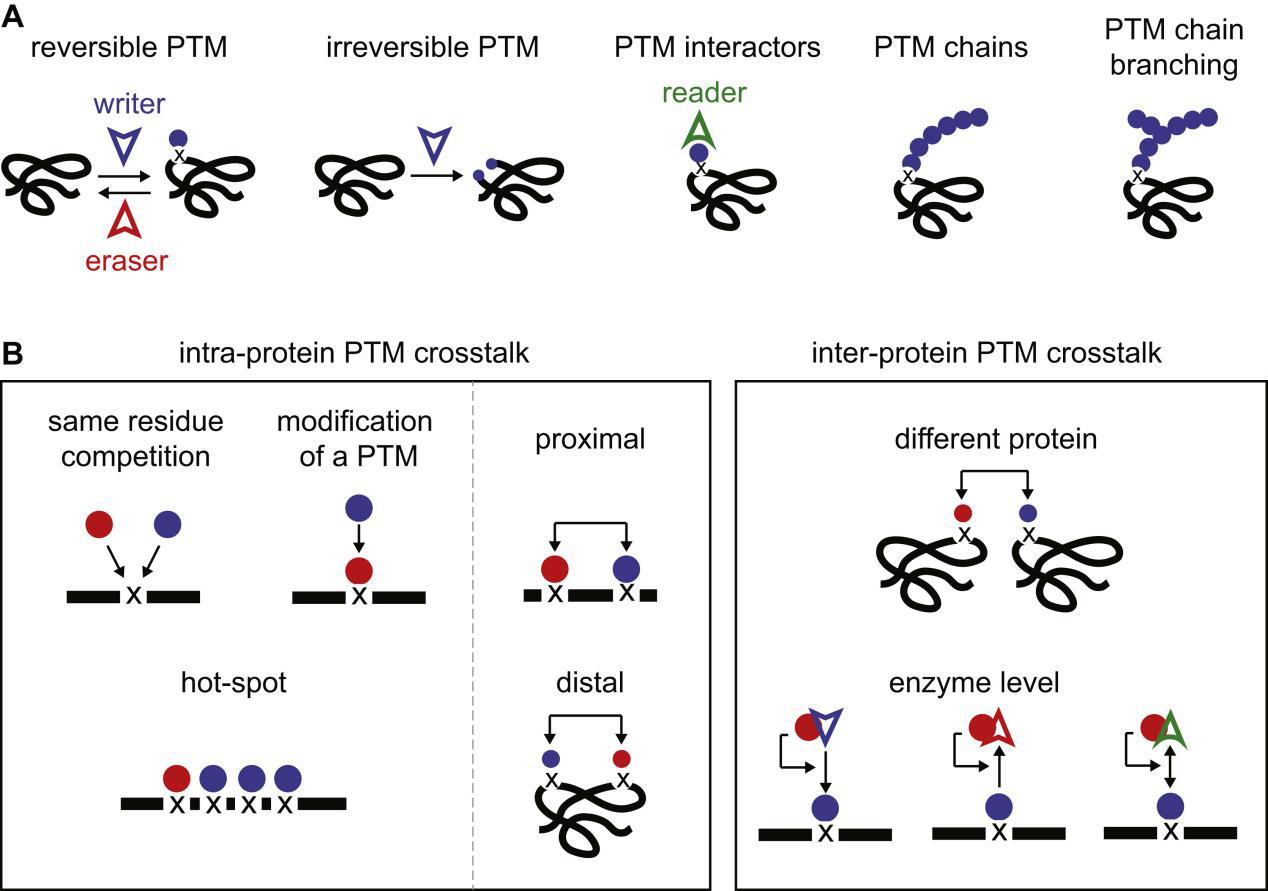
Leutert, M. et al. Mol. Cell. Proteomics. 2021.
Figure 1. PTM Crosstalk Mechanism Overview
MtoZ Biolabs, combining high-resolution mass spectrometry platforms, modification peptide enrichment technologies, and bioinformatics tools, offers the Protein PTM Crosstalk Analysis Service that supports the identification, modeling, and functional annotation of PTM crosstalk at the proteome level. This service aids in disease mechanism analysis, key target screening, and the discovery of multi-modification regulatory modes.
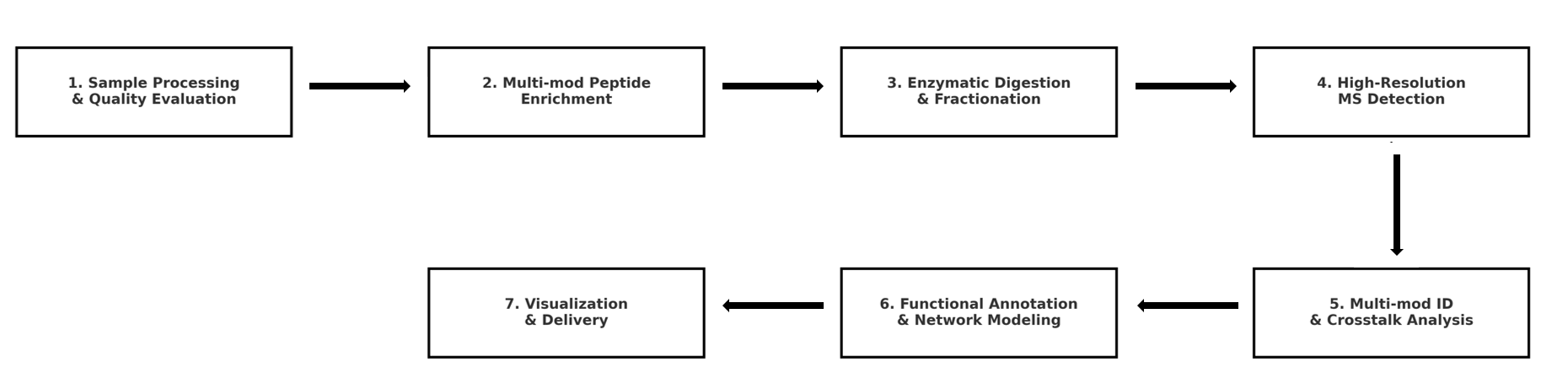
Analysis Workflow
To provide high-throughput interactive research from modification identification to functional annotation, the Protein PTM Crosstalk Analysis Service is built on a modular and standardized service process, adaptable to different sample types and research objectives. The service covers the full spectrum, from experimental detection to network modeling:
1. Sample Processing and Quality Evaluation
Our Protein PTM Crosstalk Analysis Service accepts various sample types (e.g., tissues, cells, recombinant proteins, protein complexes) with standardized preprocessing to ensure the stability of modifications during the analysis.
2. Multi-modification Peptide Enrichment Strategy
Using parallel or serial affinity enrichment methods, we design tailored enrichment strategies based on modification types, optimizing the detection efficiency of co-modified sites.
3. Enzymatic Digestion and Hierarchical Separation
Our service employs enzymatic digestion and deep peptide fractionation to enhance the detection of low-abundance modifications in complex samples.
4. High-Resolution Mass Spectrometry Detection
We utilize high-end Orbitrap systems such as Fusion, QE HF, and Lumos for Nano-LC-MS/MS detection, supporting both bottom-up and top-down strategies for comprehensive analysis.
5. Identification of Multi-modifications and Crosstalk Relationships
We identify co-modified peptides at the site level and analyze the synergy, mutual exclusivity, and sequential relationships between PTMs to construct a modification relationship matrix.
6. Functional Annotation and Network Modeling
Using integrated bioinformatics tools, we build modification–PPI (protein–protein interaction) regulatory networks, overlay GO/KEGG pathways, and analyze functional regulatory modules.
7. Results Visualization and Delivery
Finally, we provide high-quality maps and annotated tables, including interaction heatmaps, co-modification networks, and differential modification pathway diagrams, supporting publication-ready results and presentations.
Service Advantages
With our multi-dimensional integration capabilities and rich project experience, our Protein PTM Crosstalk Analysis Service offers significant advantages in the following aspects:
✅ High-Sensitivity Mass Spectrometry Support
We use Orbitrap Fusion, Fusion Lumos, QE HF, and other high-end platforms to ensure reliable detection of low-abundance modifications across multiple PTMs.
✅ Multi-modification Integration
Our service supports the integration of common PTMs such as phosphorylation, acetylation, methylation, ubiquitination, and SUMOylation.
✅ Cross-layer Modeling
We can integrate data from peptides to proteins and from proteins to networks, ensuring a comprehensive understanding of PTM crosstalk and its biological impact.
✅ Structure-guided Analysis
Our service can incorporate AlphaFold structural predictions to assist in interpreting crosstalk sites and understanding their functional implications.
✅ Multi-sample Compatibility
We adapt to a wide range of samples, including cells, tissues, protein complexes, and antibody samples.
Applications
The Protein PTM Crosstalk Analysis Service supports various research and application needs, including:
· Epigenetic modification crosstalk and chromatin state analysis
· Reconstruction of PTM co-regulatory networks in tumor microenvironment signaling pathways
· Cross-protein dependency modeling in immune protein complexes
· Building disease-specific PTM crosstalk profiles in clinical samples
· Validation and mechanism exploration of multi-modification co-regulation in drug screening
Sample Submission Suggestions
To ensure the accuracy and reproducibility of the Protein PTM Crosstalk Analysis Service, MtoZ Biolabs recommends the following sample preparation standards:
· Sample Types
Accepts tissues, cultured cells, recombinant proteins, complexes, and antibodies—suitable for various PTM-related research.
· Protein Input Requirements
≥100 µg total protein per sample; for TMT/iTRAQ quantitative analysis, ≥300 µg is recommended.
· Processing Guidelines
Use cold lysis and promptly add PTM stabilizers (e.g., NEM, IAA) to preserve native modifications and minimize artificial interference.
Need help with sample preparation? We provide detailed submission guides and personalized pre-treatment support—feel free to consult our technical team.
Deliverables
1. Experimental Overview
2. Identification & Quantification Results
3. PTM Crosstalk Analysis
4. Functional Annotation & Network Analysis
5. Data Visualization
6. Reports & Data Files
FAQ
Q1: Can the service detect patterns of multiple modifications on the same peptide? How do you ensure accurate identification?
Yes. Using high-resolution MS platforms such as Orbitrap Fusion Lumos, combined with tandem enrichment and deep fractionation strategies, we can accurately capture co-modified peptides and enhance the reliability of crosstalk identification through multiple fragmentation modes (HCD/ETD). All co-modification identifications are validated at the spectrum level to ensure data reliability and traceability.
Q2: Can you identify mutual exclusivity or synergy between different PTMs?
Yes. The Protein PTM Crosstalk Analysis Service combines statistical modeling and network co-occurrence analysis to uncover mutual exclusivity and synergy between PTMs at the same site or domain. We can also assess spatial distances between modifications using protein structure models (e.g., AlphaFold) to further verify physical exclusion or functional coupling.
MtoZ Biolabs is committed to providing high-quality, customizable protein post-translational modification analysis services. Our Protein PTM Crosstalk Analysis Service will help you gain deeper insights into the complex networks of protein modifications, accelerating disease research, therapeutic development, and biomarker discovery. For more details or project consultation, please visit our website or contact your dedicated service advisor.
Related Services
How to order?







