Protein Ligand Binding Site Detection Service
Ligand-binding site identification lies at the heart of protein function studies and drug development. These sites define where proteins specifically interact with small molecules, nucleic acids, or other proteins, and mark the starting point for key biological events like signal transduction, enzymatic activity, and molecular recognition. Pinpointing these binding regions is essential for understanding underlying molecular mechanisms, building structure-function models, and designing effective therapeutic targets.
Yet, binding sites are structurally diverse and often elusive. Some are clearly exposed on stable domains, while others are hidden within flexible regions or only become accessible upon ligand binding. They may include catalytic cores or allosteric regulatory pockets. This structural complexity poses significant challenges for comprehensive site discovery, especially in membrane proteins, large complexes, or highly dynamic systems.
To overcome these obstacles, researchers are moving beyond static structural snapshots toward integrated, multi-angle strategies. These include mass spectrometry-based techniques such as hydrogen-deuterium exchange (HDX-MS), chemical footprinting, and cross-linking MS; computational modeling and molecular docking; and conformational analysis of protein-ligand complexes. When combined, these tools offer a detailed view of how proteins engage with their ligands-linking structural features to biological function and laying the groundwork for mechanism-based drug design.
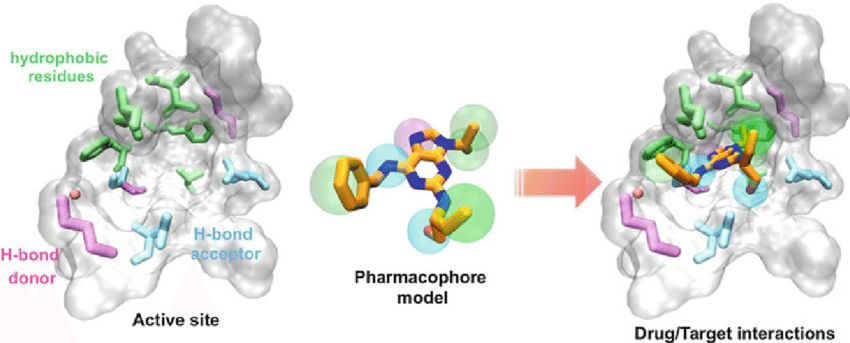
Palermo. et al. Marco. 2015.
Figure 1. Scheme of the Ligand Binding of a Small Molecule to the Target Proteins
Service at MtoZ Biolabs
MtoZ Biolabs has developed a highly integrated protein ligand binding site detection service. MtoZ Biolabs supports a wide range of target types, from single-domain proteins to transmembrane protein complexes, and handles diverse ligands including small molecules, peptides, proteins, and nucleic acids. The platform accommodates biological systems with varying structural stability and pronounced conformational dynamics.
MtoZ Biolabs integrates multiple complementary experimental strategies, combining X-ray crystallography, nuclear magnetic resonance (NMR), cryo-electron microscopy (cryo-EM), and molecular docking simulations to achieve highly sensitive detection and precise spatial localization of binding sites. MtoZ Biolabs delivers robust support for the following applications:
✅ Identifying molecular targets and characterizing small molecule binding
✅Investigating enzymatic mechanisms and locating substrate recognition regions
✅Mapping regulatory sites in proteins with dynamic conformational states
✅Defining the structural interfaces of protein-protein and protein-RNA interactions
Select Service
Protein-Ligand Structure Characterization Service
Protein Ligand Binding Assay Service
Protein-Protein Interaction Analysis Service
Ligand Receptor Analysis Service
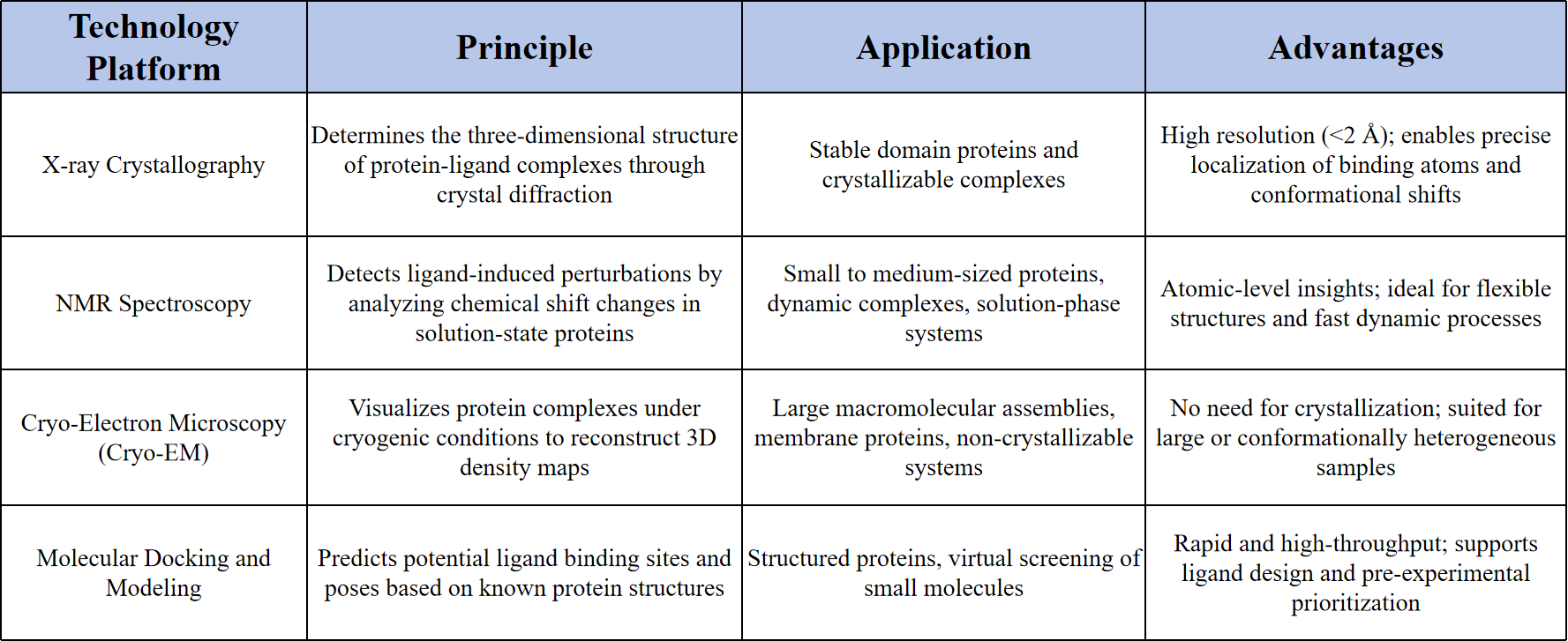
Technical Principles
Why Choose MtoZ Biolabs?
1. Multi-platform integration to accommodate diverse protein types and binding systems
MtoZ Biolabs flexibly integrates X-ray crystallography, NMR spectroscopy, cryo-EM, and molecular docking to support a wide spectrum of study targets, including stable proteins, flexible structures, membrane proteins, and large macromolecular complexes.
2. Atomic-resolution mapping for precise identification of binding interfaces
MtoZ Biolabs enables binding site localization at resolutions below 2 Å, allowing accurate identification of key residues and visualization of specific interaction patterns such as hydrogen bonds and hydrophobic contacts.
3. Support for dynamic structures and conformationally heterogeneous samples
By leveraging NMR and cryo-EM, MtoZ Biolabs analyzes dynamic binding events and induced-fit mechanisms, providing structural insights into proteins with significant conformational variability.
4. Complementary computational and experimental strategies to enhance efficiency and accuracy
MtoZ Biolabs combines molecular docking with experimental methods to predict plausible binding modes, streamline structural resolution workflows, and improve the analysis of low-affinity ligands or complexes lacking structural information.
Sample Submission Suggestions
MtoZ Biolabs supports a wide range of protein sample types for ligand binding site analysis, including but not limited to:
• Recombinant purified proteins (recommended purity ≥95% with good stability)
• Protein-ligand mixtures or pre-formed complexes
• Membrane proteins or proteins that are difficult to crystallize
• Native-source proteins
• Target proteins with known primary sequences
Recommended Sample Quantity
To ensure reliable structural analysis, MtoZ Biolabs recommends submitting at least 5-10 mg of each protein sample at a concentration of ≥1 mg/mL.
Sample Handling Guidelines
• Avoid high salt (>300 mM), glycerol, DTT, EDTA, or similar components in the buffer system. Please inform MtoZ Biolabs in advance if such components are necessary.
• Purify and store samples under sterile, protease-free conditions.
• Ensure monodispersity and absence of precipitation; size-exclusion chromatography (SEC) is recommended for purification.
• Store samples at −80°C and ship using dry ice or liquid nitrogen tanks to maintain biological activity.
For samples with complex backgrounds, poor stability, or challenging properties such as low abundance or high conformational heterogeneity, MtoZ Biolabs encourages early consultation. Our team will provide tailored recommendations to optimize analysis success.
How to order?







