Protein-Ligand Structure Characterization Service
Protein-Ligand Structure Characterization Service uses cryo-electron microscopy (Cryo-EM) to resolve the three-dimensional structures of protein-ligand complexes at high resolution without requiring crystal preparation. It enables precise modeling of binding interfaces and is suitable for various complex systems, including protein-small molecule drugs, protein-peptides, protein-DNA/RNA, and protein-cofactors.
In structural biology and drug discovery, elucidating the binding modes between proteins and ligands—such as small molecules, nucleic acids, peptides, and ions—is essential for understanding biomolecular functions, uncovering regulatory mechanisms, and advancing structure-based drug design (SBDD).
Traditionally, structural determination of protein-ligand complexes relies on X-ray crystallography (XRD) and nuclear magnetic resonance (NMR). However, these methods face significant challenges when applied to dynamic systems, difficult-to-crystallize proteins, or heterogeneous complexes. With the rapid advancement of Cryo-EM technology, structural analysis can now be performed without crystallization, while preserving near-native conformations and accommodating heterogeneous assemblies. Cryo-EM has thus become an important and innovative complementary approach for protein-ligand structure determination.
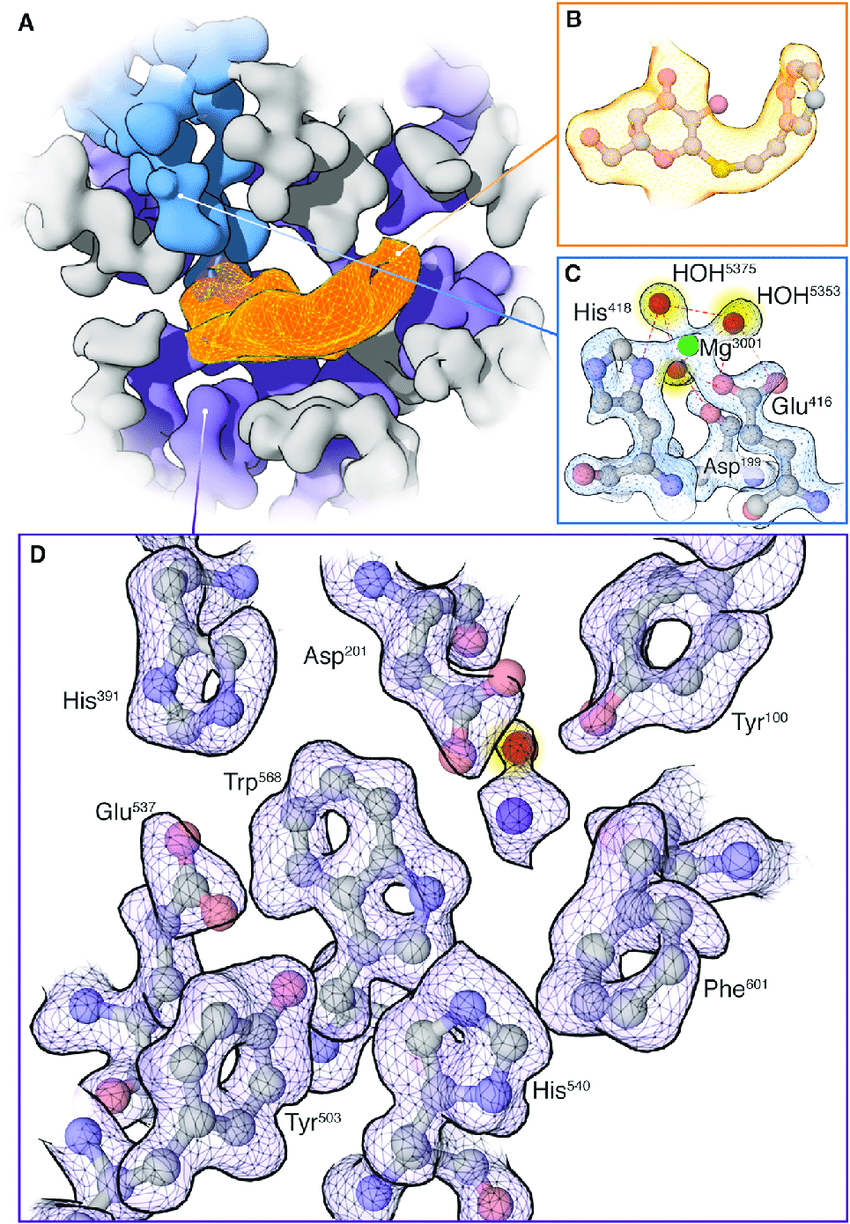
Bartesaghi A. et al. Structure. 2018.
Leveraging an advanced cryo-electron microscopy platform, MtoZ Biolabs offers the Protein-Ligand Structure Characterization Service enabling the identification of ligand-binding sites, characterization of key molecular interactions, and analysis of ligand-induced conformational changes under near-physiological conditions. This service can provide reliable support for target mechanism studies, SBDD, functional validation, and biomolecular engineering optimization.
Analysis Workflow
Protein-Ligand Structure Characterization Service provided by MtoZ Biolabs includes the following key steps:
1. Sample Preparation and Complex Assembly
Prepare high-purity protein and ligand samples; optimize complex assembly conditions to ensure binding stability.
2. Cryo-Grid Preparation and Initial Screening
Prepare cryo-EM grids and perform negative-stain TEM screening to assess complex integrity and particle distribution.
3. Cryo-EM Data Collection
Capture high-resolution images of the protein-ligand complexes in near-native states using cryo-electron microscopy.
4. Image Processing and 3D Reconstruction
Conduct particle picking, classification, and three-dimensional reconstruction to generate a 3D density map of the complex.
5. Ligand Density Localization and Structural Modeling
Identify the ligand within the density map, build atomic-level binding models, and analyze molecular interactions.
6. Structural Analysis and Data Delivery
Deliver comprehensive data packages, including 3D density maps, coordinate files, and detailed binding interface reports, suitable for scientific publication or drug development purposes.
Applications
Cryo-EM-based Protein-Ligand Structure Characterization Service is widely applicable across various life science and pharmaceutical research fields, including:
Structure-Based Drug Design (SBDD)
Resolve protein-small molecule binding modes, define binding pocket features and key interactions, and guide lead compound optimization and novel drug development.
Functional Mechanism Studies
Reveal ligand-induced conformational changes, regulation of biological activity, and catalytic mechanisms, supporting functional annotation and pathway analysis.
Protein-RNA/DNA Complex Analysis
Characterize the interaction patterns and regulatory mechanisms of transcription factors, modifying enzymes, and nucleic acid-binding proteins with RNA or DNA substrates.
Antiviral and Antibacterial Mechanism Research
Analyze binding interfaces between viral or bacterial proteins and neutralizing antibodies or small-molecule inhibitors, supporting the discovery of new anti-infective agents.
Protein Engineering and Mutation Effect Validation
Compare pre- and post-mutation binding modes to validate the effects of key residues on binding stability, affinity, and functional activity.
FAQ
Q. Can Cryo-EM reveal conformational changes induced by ligand binding?
Absolutely. Cryo-EM is highly capable of capturing both global and local conformational changes through 3D classification and local flexible modeling. It can visualize rearrangements of active sites, opening and closing of binding pockets, dimerization or oligomerization processes, and is particularly effective for capturing transient conformational states and dynamic regulatory mechanisms.
Q. Can Cryo-EM resolve structures of protein-ligand complexes with low affinity (micromolar range)?
Yes, but optimization strategies are needed, such as increasing ligand concentration, extending incubation time, using chemical cross-linking to stabilize binding, or employing local enrichment methods (e.g., affinity purification) to raise the proportion of bound complexes, thereby improving imaging and reconstruction success rates.
Q. Is Cryo-EM suitable for complexes involving multiple ligands or bridging-type small molecules?
Yes. Cryo-EM is well-suited for systems involving multiple ligands binding simultaneously (e.g., bivalent small molecules or bridging complexes). It can use conformational classification to distinguish different binding modes and resolve the spatial organization and interaction mechanisms of each binding site.
Case Study
This study used Cryo-EM technology to analyze the three-dimensional structure of tau protein paired helical filaments (PHFs) extracted from the brains of Alzheimer's patients after binding to the PET imaging ligand MK-6240. The results showed that MK-6240 bound to the C-shaped cavity inside PHF in a 1:1 ratio, mainly interacting with residues such as Gln351, Lys353 and Ile360, and stacking obliquely along the fiber axis to establish a stable π-π interaction network. This study revealed for the first time the specific sites and patterns of MK-6240 binding to tau fibers at atomic resolution, providing an important structural basis for understanding its imaging specificity, optimizing PET ligand design, and studying the pathological mechanisms of Alzheimer's disease.
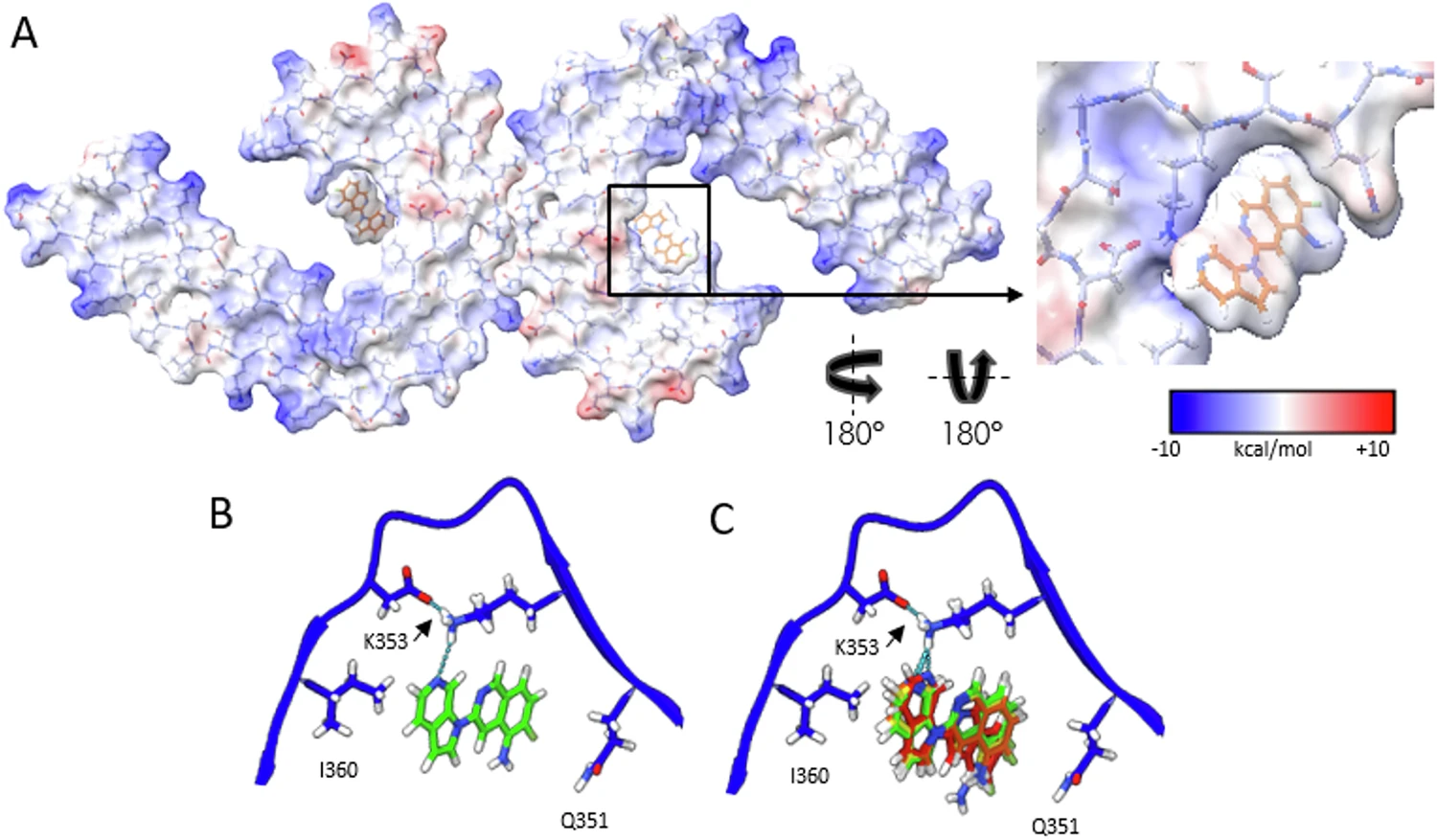
Kunach P. et al. Nat Commun. 2024.
How to order?







