Protein Full Sequence Coverage Analysis Service
The analysis of protein full sequence is a critical and challenging endeavor, especially against the backdrop of rapid advancements in the biopharmaceutical industry. Sequence verification has become a cornerstone for the development of biosimilars and biologic therapeutics. Furthermore, the quest for deeper insights into the molecular mechanisms of biological processes and disease initiation amplifies the need for more efficient and precise protein sequencing technologies. Complete protein sequence information is imperative for confirming the intact expression of recombinant proteins, identifying potential sequence breakages or mutations, and providing vital metrics for biomarker discovery, disease diagnosis, drug development, and personalized medicine.
MtoZ Biolabs offers an exhaustive service for protein full sequence coverage analysis. Our team is adept at determining the complete sequence coverage of recombinant proteins or other target proteins from any species, conformation, and state, including samples derived from diseased tissues and recombinantly expressed proteins such as protein pharmaceuticals, antibody drugs, vaccines, peptide drugs, and collagen.
MtoZ Biolabs utilizes multi-enzyme digestion and combined enzyme digestion pre-treatment methods to ensure a comprehensive and diverse collection of target protein peptide sequences. By employing a high-resolution mass spectrometry analysis platform, we have developed a technique centered around mass spectrometry for in-depth protein full sequence analysis, capable of achieving 100% coverage of target protein sequences.
High-Resolution Mass Spectrometry
High-resolution mass spectrometry stands as a sophisticated tool for analyzing chemical and biological molecules, offering precise insights into molecular structures and compositions. Its application spans across chemistry, biology, pharmaceutical research, environmental science, and food safety. This technology precisely determines the molecular mass of analytes based on the charge-to-mass ratio of charged ions, and in tandem mass spectrometry, allows analytes to further fragment, producing a spectrum of fragment ions. Utilizing high-resolution mass spectrometry enables the precise determination of the amino acid sequence of protein-specific enzymatic digests and, through sequence assembly, completes the protein sequence coverage analysis.
Experimental Instruments
1. Liquid Chromatography System (Easy-nLC 1200)
2. Electrospray Ionization Hybrid Orbitrap Mass Spectrometer (Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer, Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer)
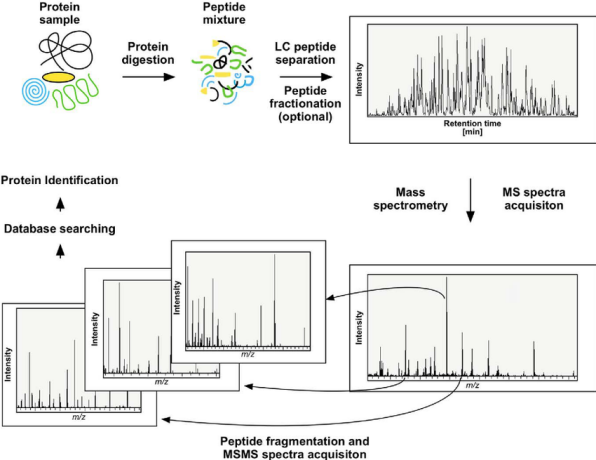
Figure 1. Principles of Protein Sequence Identification via High-Resolution MS
Analysis Workflow
1. Enzymatic Digestion of the Target Protein Using Specific Proteases
2. Mass Spectrometry Analysis of the Digested Sample
3. Sequence Identification Using Professional Analysis Software
4. Assembly of the Sequence Identification Results of Multiple Eenzyme Digestions to Obtain Complete Protein Sequence Information
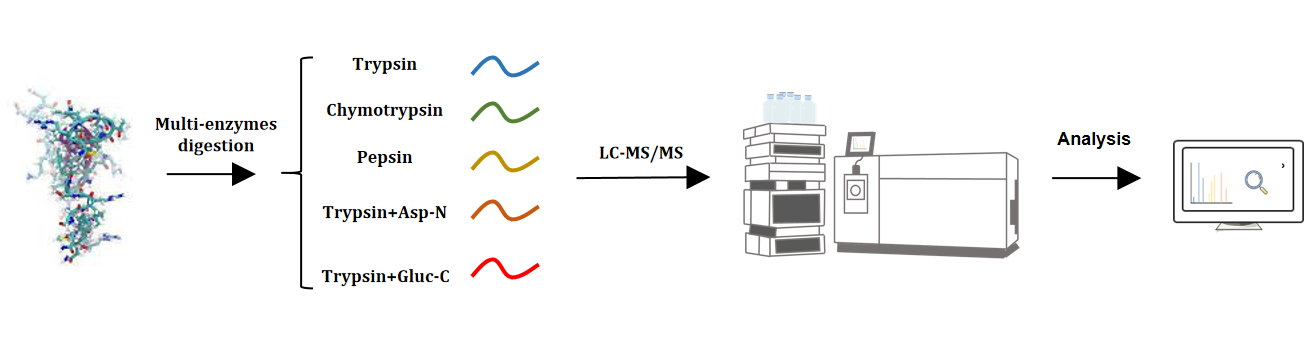
Figure 2. Achieving Comprehensive Protein Sequence Coverage Analysis Through Multiple Enzymatic Digestion Strategies
Service Advantages
1. Employment of various enzymatic digestions, such as Trypsin, Chymotrypsin, Asp-N, Pepsin, Glu-C, and combined digestions (e.g., Trypsin+Asp-N, Trypsin+Glu-C), for target protein digestion and identification.
2. Use of high-resolution mass spectrometry and professional identification software to ensure result accuracy.
3. While obtaining the coverage of single enzyme digestion, the results of each enzyme digestion were combined to obtain the full sequence information of the protein.
4. Verification of protein sequence completeness and detection of potential site mutations or peptide cleavages.
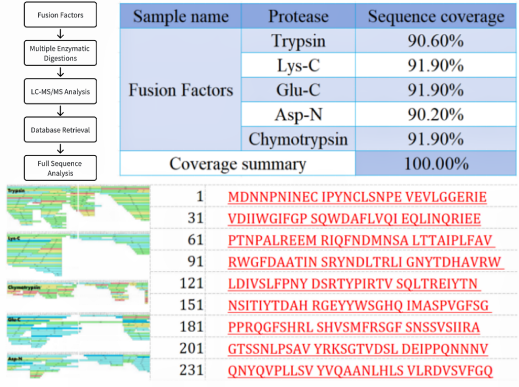
Sample Results
Sample Submission Requirements
1. Both gel and solution samples are acceptable for the sequencing process.
2. Required target protein sample amount: 50-100 μg.
3. To ensure accurate detection of the target protein coverage rate, maximize sample purity.
4. Analyzed sample types include protein pharmaceuticals, antibody drugs, vaccines, peptide drugs, and recombinant collagen.
Services at MtoZ Biolabs
Through our protein full sequence coverage analysis service, you will obtain detailed coverage under each enzymatic condition and the complete protein coverage from various enzymatic digestions, providing a standard for assessing your sample's sequence integrity. We also provide related experimental steps, mass spectrometry parameters, total ion chromatograms, original mass spectrometry data, and detailed protein full sequence coverage information.
Applications
1. Verification of recombinant protein sequences.
2. Determination of the accuracy of amino acid full sequences in proteins, peptide drugs, antibodies, and vaccines.
3. Consistency of sequences expressed under different batches, experimental methods, and conditions.
4. Clinical biomarker sequence verification.
FAQ
Q1: What does coverage indicate? What should be done if the coverage is not 100%?
Coverage refers to the proportion of sequenced sequences to the entire sequence, reflecting the extent to which our sequencing results encompass the target region. If the coverage rate is 100%, it implies that the sequence obtained after enzymatic digestion closely matches the target sequence. If the coverage is less than 100%, we will review the protein data and sequence to ascertain the extent of sequence loss and identify undetected theoretical peptides. Decisions on whether to continue with current methods and enzymatic treatments to recover missing peptide locations will be based on these findings.
Q2: How should I prepare a protein sample with a small molecular weight for sequencing?
For proteins with small molecular weights, utilize high-concentration SDS-PAGE to separate the proteins, followed by Coomassie staining of the gel. Cut out the protein bands corresponding to the target protein for mass spectrometry identification.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







