Protein Disulfide Bonds Identification and Quantitative Analysis Service
Protein Disulfide Bonds Identification and Quantitative Analysis Service is a service that identifies and quantifies disulfide bonds in protein molecules. The three-dimensional structure of proteins is the basis for the function of proteins. In many proteins, disulfide bonds are key covalent bonds that maintain their three-dimensional structure. Disulfide bonds are covalent bonds formed by sulfur atoms between two cysteine residues through oxidation reactions. They are widely present in many proteins, especially in higher-order structures. They play a vital role in the stability and function of proteins, especially in cell signaling, protein complex formation, and enzyme catalysis. The analysis of protein disulfide bonds not only helps to deeply understand the three-dimensional structure and biological function of proteins, but is also crucial for protein engineering, drug development, and the study of disease mechanisms.
Relying on advanced mass spectrometry platforms, MtoZ Biolabs provides Protein Disulfide Bonds Identification and Quantitative Analysis Service to accurately identify disulfide bonds in proteins and quantify their dynamic changes under different conditions, providing researchers with information about protein structure, function and its changes under different physiological or pathological states.
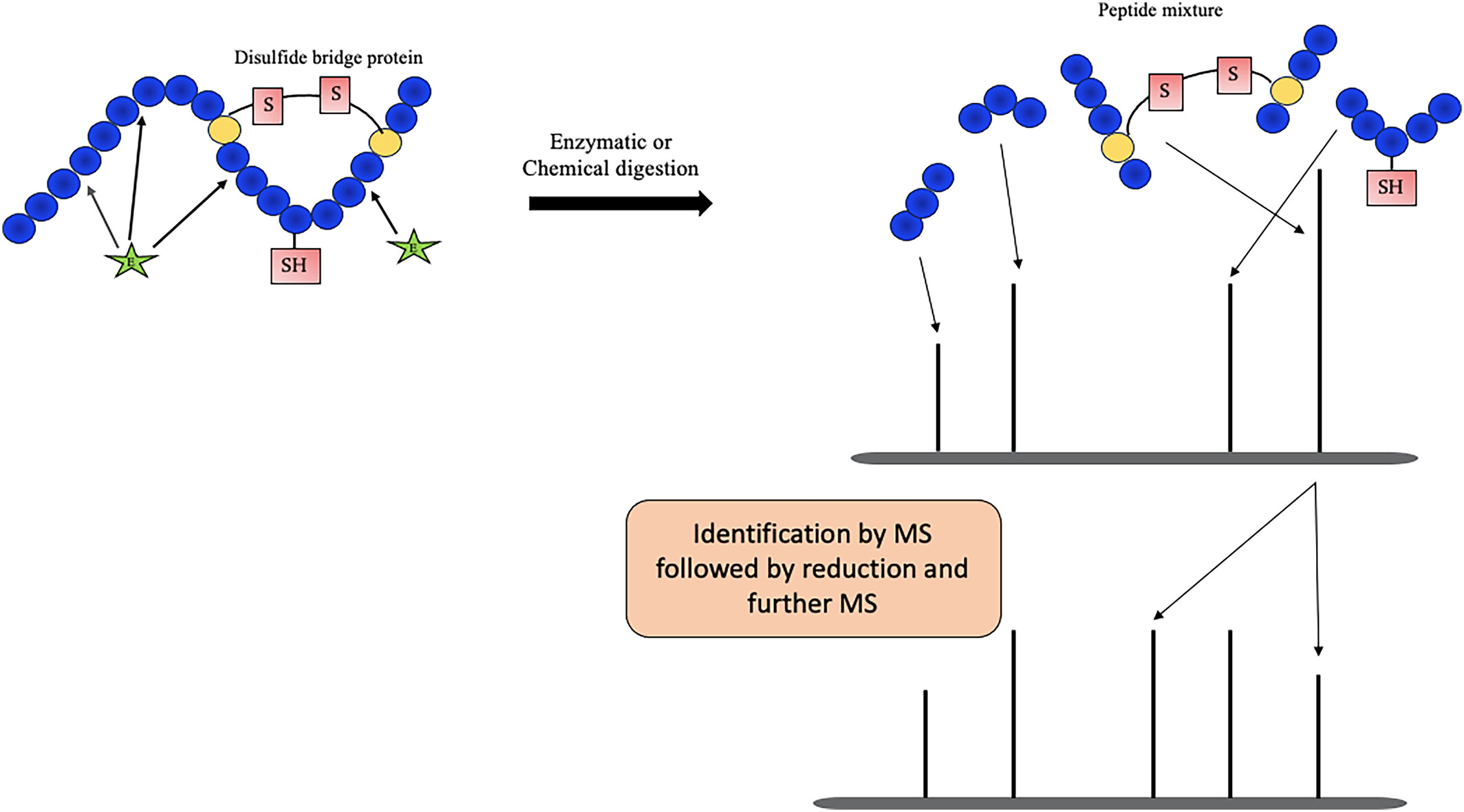
Chaturvedi S. et al. Rapid Commun Mass Sp. 2024.
Figure 1. Mass spectrometry analysis of disulfide bonds.
Applications
Protein structure and function research
Protein Disulfide Bonds Identification and Quantitative Analysis can help reveal how disulfide bonds affect the three-dimensional structure and function of proteins.
Quality control of biological preparations
In the development and production of biological drugs, the analysis of disulfide bonds helps to ensure the correct folding and function of protein drugs.
Disease research
Study diseases related to abnormal disulfide bonds, such as cancer, Alzheimer's disease, etc., to reveal their potential role in disease mechanisms.
Protein engineering and optimization:
In protein engineering, the performance of recombinant proteins or enzymes is optimized by analyzing the formation and changes of disulfide bonds.
Services at MtoZ Biolabs
MtoZ Biolabs provides Protein Disulfide Bonds Identification and Quantitative Analysis Service including protein disulfide bond analysis at the single protein and omics levels. We have established sample preparation methods to overcome the in vitro exchange of protein disulfide bonds and maintain their native structure, mass spectrometry analysis methods for disulfide bond cross-linked peptides, and supporting disulfide bond identification software. MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption.
FAQ
Q. How to optimize mass spectrometry for the detection of low-abundance disulfide peptides?
To improve the detection sensitivity of low-abundance disulfide peptides, optimization can be performed in the following ways:
1. Enrichment strategy: Use antibody enrichment or chemical probes to capture specific disulfide-modified peptides, which can increase the signal intensity of the target modification and reduce background noise.
2. Liquid chromatography (LC) optimization: By optimizing the gradient and flow rate of liquid chromatography, the separation effect of peptides can be improved and signal loss can be reduced.
3. Mass spectrometry parameter optimization: Using a high-resolution mass spectrometer (such as the Orbitrap series) combined with fragmentation modes suitable for modified peptides such as HCD (high-energy collision dissociation) or ETD (electron transfer dissociation) can help improve the signal intensity and detection accuracy of low-abundance disulfide peptides.
Through these optimizations, the detection ability of low-abundance disulfide peptides can be improved and the accurate identification of disulfide bonds can be ensured.
Q. How to quantify the abundance of disulfide bonds and evaluate their changes under different conditions?
The quantification of disulfide bond abundance is usually achieved through the intensity of mass spectrometry signals. The specific steps are as follows:
1. Relationship between signal intensity and abundance: The intensity of mass spectrometry signals is usually proportional to the abundance of peptides, so the abundance of disulfide bond peptides can be inferred by measuring the peak area or peak height of disulfide bond peptides.
2. Standardization and quantitative methods: Labeling quantitative methods (such as TMT) can be used to label samples to reduce sample variation and improve quantitative accuracy.
3. Comparison of changes under different conditions: By comparing disulfide bond peptides in the experimental group with the control group or samples at different time points, the changes of disulfide bonds under different conditions can be evaluated.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study used high-resolution mass spectrometry and chemical modification to identify abnormal disulfide bonds formed on actin under sulfidation stress, and quantitatively analyzed the changes in the abundance of these disulfide bonds under different experimental conditions. The results showed that sulfidation stress significantly increased disulfide bond modifications at specific sites in actin, leading to skeleton disintegration and instability. Ultimately, these changes triggered a new mode of cell death - disulfidptosis, indicating that disulfide bond formation in the actin skeleton plays a key role in this cell death mechanism.
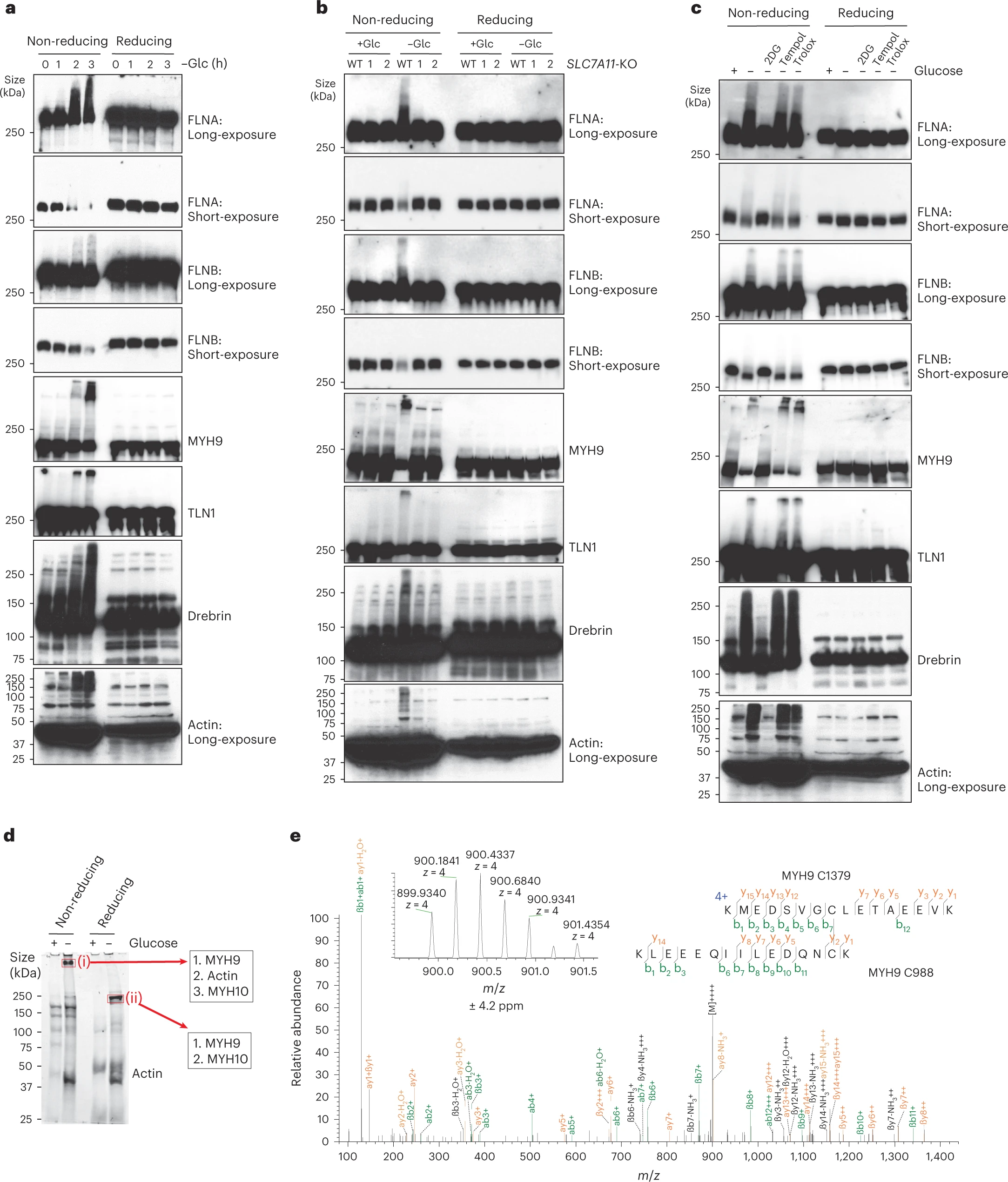
Liu X. et al. Nat Cell Biol. 2023.
Figure 2. Disulfide bonding in cytoskeleton proteins in disulfidptosis.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Biopharmaceutical Disulfide Bond Analysis Service
How to order?







