Protein Complementation Assay Service
Protein complementation assay (PCA) is a highly sensitive and quantitative high-throughput screening method that allows dynamic observation of protein-protein interactions at the level of live cells, organs, or even entire organisms. It has been widely applied in various research fields, including screening interacting proteins, drug delivery, chemical genetics, and proteomics. Using the principle of reconstitution of reporter protein fragments, MtoZ Biolabs provides efficient and precise protein complementation assay service for real-time detection and analysis of the dynamic processes of protein-protein interactions. This service helps researchers uncover key protein interaction networks in natural physiological environments, explore signaling pathway regulatory mechanisms, and assist in drug target screening and validation.
Proteins cannot function alone and rely on protein-protein interactions to function. Protein-protein interactions (PPI) are a critical component of cellular biochemical reaction networks and play a key role in discovering, identifying, and functionally validating new proteins and related protein complexes, as well as in the study of cellular signal transduction and metabolism. PPIs have become an important area in proteomics research.
To date, several methods have been used to study protein-protein interactions, including yeast two-hybrid technology, co-immunoprecipitation, immunofluorescence co-localization, pull-down assays, affinity chromatography-mass spectrometry, fluorescence resonance energy transfer (FRET), and bimolecular fluorescence complementation (BiFC). These techniques have revealed numerous complex protein interactions within cells. Among them, the PCA, which has emerged more recently, is gaining increasing attention because it allows dynamic studies of protein-protein interactions both in vivo and in vitro. Currently, PCA systems based on reporter proteins such as dihydrofolate reductase, β-lactamase, luciferase, and fluorescent proteins have been developed.
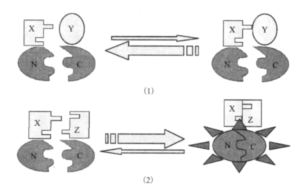
Zhang, jie. et al. Biotechnology Bulletin, 2013.
Figure 1. PCA Principle Illustrated Using Fluorescent Proteins.
Services at MtoZ Biolabs
There are several methods for protein interaction analysis, and MtoZ Biolabs offers protein interaction analysis services, along with subsequent LC-MS/MS-based identification and analysis of protein mixtures in purified samples from IP, Co-IP, and GST fusion protein pull-down assays. Our services cover the following core technical directions:
1. PCA for Interaction Analysis
The core principle of the PCA is to divide a reporter protein into two non-functional fragments, which are each fused with the two proteins of interest (Protein A and Protein B). When these proteins interact physically, either in vivo or in vitro, the two fragments are brought into close proximity and reassemble into a functional, complete reporter protein. This restores its activity, generating a detectable signal (such as fluorescence, luminescence, or enzymatic activity). PCA provides an effective tool for directly detecting protein-protein interactions, enabling not only the detection of the interaction's presence but also the real-time monitoring of dynamic interaction changes.
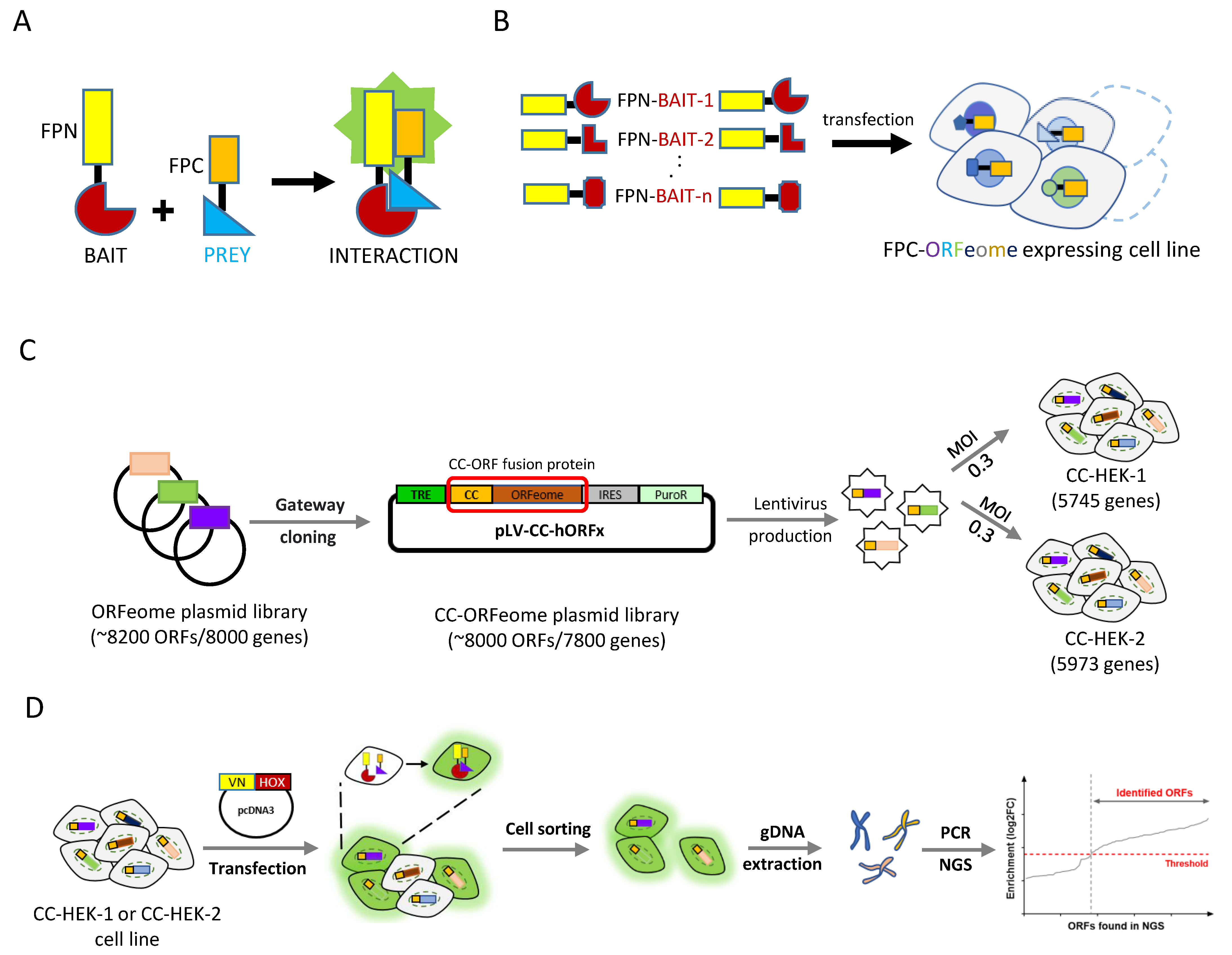
Jia, Y. et al. Cells, 2023.
Figure 2. Application of PCA-based strategies for large-scale interaction screening in living cells.
Leveraging an advanced PCA platform and an experienced research team, MtoZ Biolabs offers high-quality and cost-effective protein complementation assay services to facilitate the detection of protein-protein interactions. Based on different research needs, we flexibly choose fluorescent proteins or luminescent proteins as reporter genes to enable in vivo imaging and real-time dynamic monitoring. Additionally, MtoZ Biolabs provides extended technologies such as Bimolecular Fluorescence Complementation Resonance Energy Transfer (BiFC-RET) to assist customers in conducting more comprehensive and in-depth protein interaction studies. In experimental design, we carefully consider key factors such as reporter gene selection, reporter fragment characteristics, linker design, protein fusion topology, expression systems, and experimental controls to ensure high reliability and reproducibility of the results. This provides strong support for protein interaction mechanism analysis and drug development.
2. Co-Immunoprecipitation (Co-IP) Protein Interaction Analysis
Co-Immunoprecipitation (Co-IP) is a method that uses the specificity between an antigen and antibody to detect physiological interactions between proteins. In this experiment, non-ionic detergents are used to lyse cells to preserve protein interactions as much as possible. When the cells are lysed, the protein interactions in the cells are preserved, and the corresponding antibodies are added to bind the antibodies to the specific proteins in the cells. Proteins A/G are then added to form antibody-protein complexes, which are then centrifuged and separated. The antibody-protein complex is precipitated, the supernatant is discarded, and the precipitated protein is finally identified using mass spectrometry to prove the interaction between the two. This method is suitable for analyzing stable or strong protein-protein interactions and is typically used to identify interactions between a target protein and another protein or to discover unknown proteins that interact with a specific target protein.
MtoZ Biolabs provides optimized solutions for protein extraction, expression, and purification, ensuring high-quality bait and prey proteins. We offer excellent Co-IP protein interaction analysis services and mass spectrometry analysis of proteins obtained from Co-IP experiments.
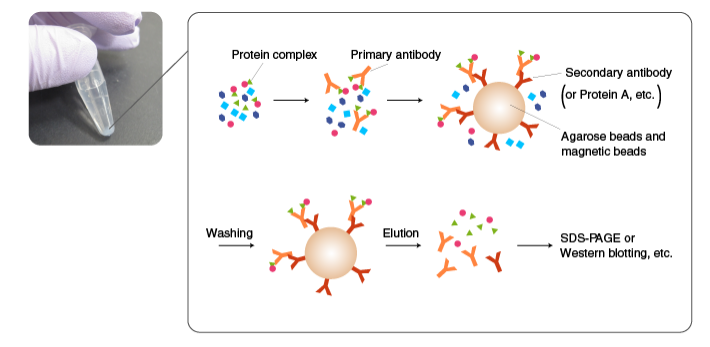
Halo, Y. et al. Journal of Immunology Research & Reports, 2021.
Figure 3. Protein interaction analysis process.
3. GST Fusion Protein Pull-Down Technology for Protein Interaction Analysis
GST Fusion Protein Pull-Down technology utilizes recombinant genes to insert a GST-tagged vector into the target protein A. The glutathione-coated magnetic beads bind with the target fusion protein. After adding cell lysate, interacting proteins are adsorbed by the fusion protein. Excess glutathione is added to elute the proteins, which are then analyzed using mass spectrometry (MS). This method primarily investigates strong or stable protein-protein interactions in vitro. It can identify direct interactions between two known proteins or discover unknown proteins that may interact with a target protein.
As a technology-driven research service company, MtoZ Biolabs has years of experience in protein interaction analysis services and mass spectrometry-based protein interaction identification. We offer GST pull-down protein interaction analysis services and pull-down target protein mass spectrometry identification services. Our strict quality control and short turnaround times provide customers with customizable, one-stop services covering every step of the project, from gene synthesis to data reporting.
4. SILAC and Co-Immunoprecipitation Mass Spectrometry for Protein Interaction Analysis
Using the SILAC method, experimental and control cells are labeled separately, followed by Co-IP experiments. Immunocomplexes are isolated based on the specificity of the antigen-antibody interaction, and proteins in these complexes are qualitatively and quantitatively detected using LC-MS/MS. When the protein content in the experimental group differs significantly from that in the control group, it is concluded that this protein interacts with the protein being studied. This method significantly reduces the possibility of false positive protein-protein interaction results. The combination of SILAC technology and Co-IP-MS can be used for high-throughput, quantitative analysis of protein interaction networks under specific conditions.
MtoZ Biolabs uses Thermo Fisher’s Q Exactive HF, Orbitrap Fusion, and Orbitrap Fusion Lumos mass spectrometers coupled with Nano-LC to offer SILAC-based Co-IP-MS protein interaction analysis services. Customers only need to provide the experimental goals and cell samples, and we will handle all aspects of the project, including cell culture, labeling, protein extraction, antibody IP, efficiency testing, protease digestion, peptide separation, mass spectrometry analysis, raw data analysis, and bioinformatics analysis.
Service Advantages
1. Advanced Analytical Platform
MtoZ Biolabs has established an advanced PCA service platform, ensuring reliable, fast, and high-precision analysis services.
2. Transparent Pricing
Our pricing is transparent, with no hidden or additional fees.
3. High-Quality Data
We offer deep data coverage and strict quality control. Our AI-driven bioinformatics platform integrates all data from PCA service to provide comprehensive reports.
4. Customized Research Solutions
MtoZ Biolabs provides tailored services to address your unique research questions and experimental requirements.
Applications
1. Screening Interaction Protein Libraries
The simplicity, sensitivity, and robustness of in vitro β-lactamase PCA provide a useful method for large-scale protein interaction analysis.
2. cDNA Library Screening
It helps identify and verify the role of proteins in any cellular process and identify enzyme substrates and regulatory proteins.
3.Biochemical Network Mapping
It enables the creation of dynamic maps and identification of new components that may play roles in the network. This method improves accuracy compared to traditional targeted drug therapies.
4. Receptor-Ligand Interaction and Potential Therapeutics
Development of imaging techniques for quantifying receptor-ligand complexes under physiological conditions has accelerated research in disease mechanisms and drug delivery.
5. Protein Detection and Subcellular Localization
The PCA service enables protein detection and subcellular localization.
6. Screening Specific Antibodies
The protein complementation assay service system is used to screen antibodies, offering high specificity, efficiency, and simplicity, with no need for antigen purification and immobilization—just cloning, transformation, and plating.
7. Screening Protein-Protein Interaction Inhibitors
The resulting reporter protein activity signals will be higher or lower depending on the presence or absence of interference factors, making the protein complementation assay service ideal for screening PPI activators or inhibitors and studying the impact of drugs on PPI, an essential tool for drug discovery.
8. Protein Interaction Network Mapping
The PCA enriches interaction network maps, providing insights into critical cellular processes such as cell polarization and autophagy.
Case Study
1. New in vitro protein complementation test system suitable for 1536-well high-throughput screening
Protein-protein interaction (PPI) plays a crucial role in all cellular processes and can serve as potential targets for new drug development. To facilitate screening of PPI inhibitors as anticancer drugs, the authors developed a high-throughput screening system using the monomeric Kusabira-Green fluorescent protein (mKG) for in vitro PCA. The in vitro PCA service system is based on the fusion of two split inactive mKG fragments with the target protein to form a functional complex. When the two target proteins interact, they emit fluorescence, thereby achieving the complementation of the mKG fragments. In this study, the authors developed an HTS-compatible in vitro PCA service system using mKG, which is easy to handle, simple to process, and does not require protein purification or refolding.
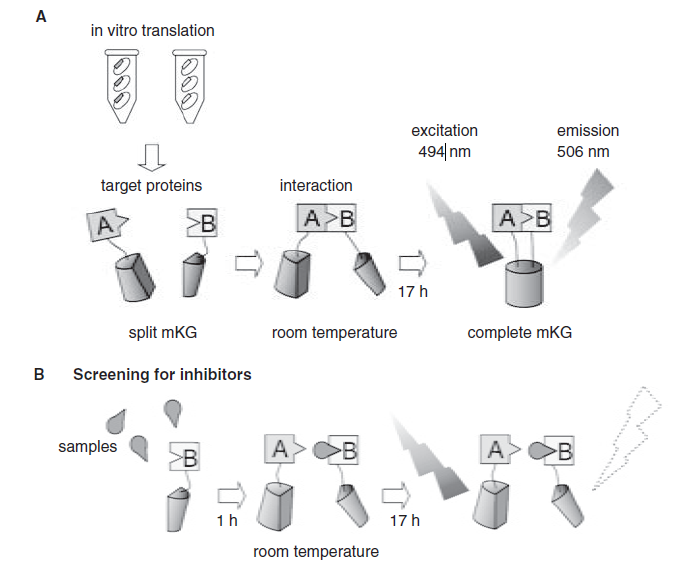
Hashimoto J. et al. Journal of Biomolecular Screening, 2009.
Figure 4. Schematic diagram of protein complementation assay.
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Co-Immunoprecipitation Protein Interaction Analysis Service
How to order?







