Post-Translational Modification Functional Analysis Service
- Revealing the mechanistic roles of post-translational modifications in signaling pathway regulation.
- Identifying disease-associated key modified proteins and their regulatory networks.
- Supporting drug target screening and mechanism validation.
- Analyzing the impact of coordinated regulation by multiple PTMs on cellular functions.
- Facilitating functional annotation and pathway association analysis in systems biology and integrative omics research.
Post-Translational Modification Functional Analysis Service refers to a professional service that utilizes high-throughput mass spectrometry data combined with systematic bioinformatics tools to deeply analyze the specific functional roles of post-translational modifications (PTMs) in cellular processes. Through pathway enrichment analysis, protein–protein interaction (PPI) network construction, functional annotation, and upstream regulator prediction, this service comprehensively deciphers the functional relevance of PTMs in signal transduction, biological regulation, and disease mechanisms.
PTMs play a central role in regulating cellular activities, influencing protein stability, activity, subcellular localization, and signal transduction. Systematic functional analysis of PTMs is essential for understanding cell fate decisions and disease pathogenesis. PTM functional analysis employs comprehensive strategies to biologically interpret the functional changes induced by specific modifications on proteins.
This type of analysis is typically based on high-throughput mass spectrometry identification results, supported by advanced bioinformatics tools such as Gene Ontology (GO) annotation, KEGG pathway enrichment analysis, protein–protein interaction (PPI) network construction, motif identification, and upstream kinase prediction. These approaches help identify key modified proteins and their involvement in core regulatory processes, enabling a functional understanding of PTM-driven mechanisms.
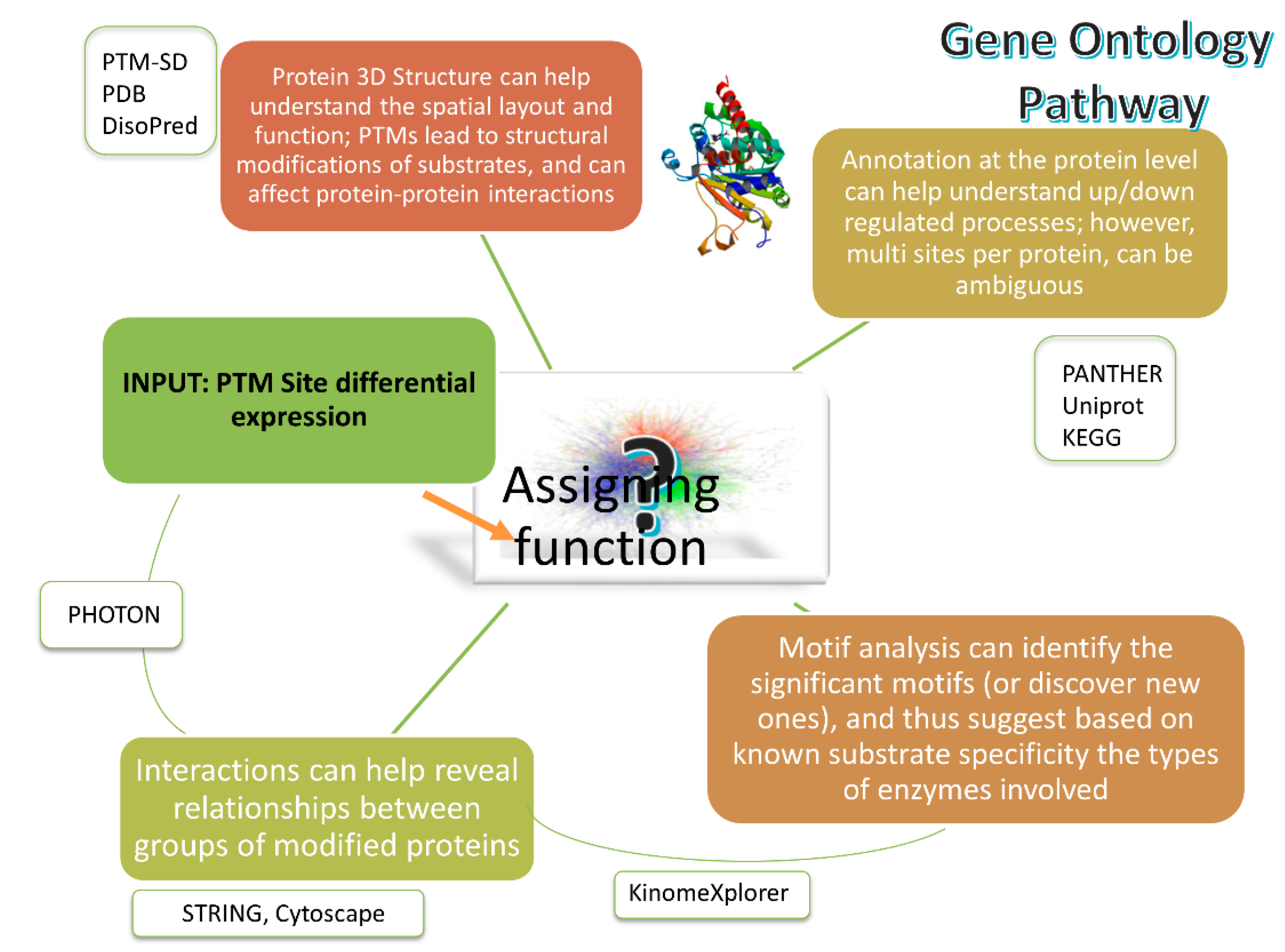
Pascovici D. et al. Int J Mol Sci. 2019.
Leveraging advanced bioinformatics analysis methods, MtoZ Biolabs provides Post-Translational Modification Functional Analysis Service to deeply investigate the functional roles of protein PTMs in biological processes, facilitating comprehensive translation from data to mechanistic insights. In addition, MtoZ Biolabs also offers high-resolution mass spectrometry-based services for PTM site identification and quantification as a foundation for functional analysis.
Analysis Workflow
The main workflow of Post-Translational Modification Functional Analysis Service is as follows:
1. Input Data Processing
Clients may submit PTM-modified protein data, or MtoZ Biolabs can generate the data via PTM analysis. All input is validated and preprocessed for downstream analysis.
2. Functional Annotation and Pathway Enrichment
Annotate modified proteins using multiple databases and assess functional significance through enrichment analysis.
3. Protein–Protein Interaction (PPI) Network Construction
Build interaction networks among modified proteins to identify potential regulatory hubs.
4. Optional Module Analyses
Support additional analyses such as motif identification, regulatory enzyme prediction, and structural or visual interpretation.
5. Results Integration and Report Delivery
Provide a comprehensive analysis report with figures, tables, and raw data, along with optional customized interpretation services.
Service Advantages
1. High Compatibility with Input Data: Supports data from various mass spectrometry platforms.
2. Comprehensive Database Integration: Full coverage of high-quality resources such as GO, KEGG, and STRING.
3. Modular Analytical Framework: Flexible selection of core functional analysis and advanced regulatory modules based on research objectives.
4. High-Quality Visual Outputs: Delivers diverse visualizations—including enrichment plots, network diagrams, and heatmaps—that meet publication standards.
Sample Submission Suggestions
Data Type: Protein post-translational modification data obtained from mass spectrometry analysis.
Data Format: Standard proteomics output is recommended, including protein IDs, modification types, modification sites, and quantitative information (if available).
Additional Notes: If no existing data is available, MtoZ Biolabs can provide comprehensive services from sample preparation to mass spectrometry analysis.
Applications
Typical applications of Post-Translational Modification Functional Analysis Service include:
Deliverables
1. GO functional annotation results of modified proteins
2. Pathway enrichment analysis report
3. Protein–protein interaction (PPI) network diagram
4. Optional motif and structural visualization plots
5. Editable-format analytical charts and raw data files
Related Services
Post-Translational Modification Site Motif Analysis Service
Post-Translational Modification Site Prediction Service
How to order?







