Phosphoproteomics Analysis Service
Phosphoproteomics Analysis Service utilizes high-resolution mass spectrometry (MS) technology to systematically identify, quantify, and functionally analyze phosphorylated proteins in biological samples. Phosphorylation is one of the most critical post-translational modifications, playing a key role in cellular signal transduction, cell cycle regulation, metabolic control, immune response, and disease progression. This modification is typically catalyzed by kinases and can be reversibly regulated by phosphatases, dynamically controlling protein activity, stability, localization, and interactions. Phosphoproteomics Analysis Service not only enables comprehensive identification of phosphorylation sites but also tracks dynamic changes in phosphorylation levels, uncovering kinase-substrate interactions and regulatory mechanisms of signaling pathways. Leveraging a high-resolution mass spectrometry platform combined with highly efficient phosphopeptide enrichment strategies, MtoZ Biolabs provides an end-to-end phosphoproteomics analysis service from sample preparation to data interpretation ensuring high-confidence results for clients.
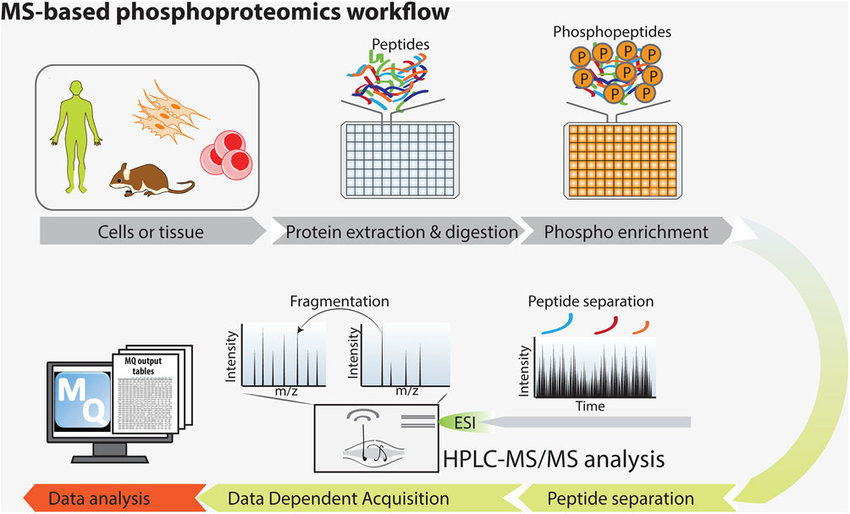
Sacco. et al. Proteomics. 2018.
Analysis Workflow
1. Sample Preparation
Protein extraction and enzymatic digestion. Standard trypsin or Lys-C is used for digestion, with optional phosphatase inhibitor treatment to prevent the loss of phosphorylation signals in vitro.
2. Phosphopeptide Enrichment
Phosphopeptides are enriched using strategies such as TiO₂, IMAC, or Fe-NTA. High-performance liquid chromatography (HPLC) is applied for pre-fractionation to reduce non-specific binding and enhance data quality.
3. Mass Spectrometry Data Acquisition
High-resolution mass spectrometry is used for analysis. Data acquisition strategies include Data-Independent Acquisition (DIA), Data-Dependent Acquisition (DDA), and Parallel Reaction Monitoring (PRM). Quantitative analysis of phosphorylated proteins is performed using Label-Free, TMT/iTRAQ, or other labeling strategies.
4. Data Analysis
Phosphorylation Site Identification: Peptide identification is conducted using specialized software.
Quantitative Analysis: Quantification of phosphorylated proteins across different samples, with statistical significance analysis.
Bioinformatics Analysis: Functional enrichment analysis (GO, KEGG), kinase-substrate analysis, and protein-protein interaction (PPI) network analysis are performed.
Applications
Some applications of the Phosphoproteomics Analysis Service are as follows:
Cell Signaling Pathway Research
Unravel the phosphorylation regulatory mechanisms in key signaling pathways to reveal how cells respond to external stimuli.
Disease Research & Biomarker Discovery
Identify disease-associated phosphorylated proteins, screen for potential biomarkers, and investigate disease progression and drug resistance mechanisms.
Drug Development & Target Screening
Evaluate the mechanism of action of kinase inhibitors and identify potential drug targets.
Immune & Metabolic Regulation
Study phosphorylation-mediated regulation in T-cell activation and B-cell receptor signaling pathways. Analyze the role of phosphorylated metabolic enzymes in glucose metabolism and lipid metabolism.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs offers Phosphoproteomics Analysis Service covering phosphoprotein identification, phosphorylation site mapping, and quantitative phosphoproteomics. We provide customized solutions tailored to your specific research needs. Feel free to contact us for consultation.
FAQ
Q. What is the optimal method for phosphopeptide enrichment?
The selection of a phosphopeptide enrichment method depends on factors such as sample type, phosphorylation level, and desired depth of analysis. Below are the characteristics and suitable applications of different enrichment strategies:
IMAC (Immobilized Metal Affinity Chromatography): Capable of enriching both Ser/Thr- and Tyr-phosphorylated peptides, making it suitable for large-scale phosphoproteomics studies.
TiO₂ (Titanium Dioxide): Highly specific for Ser/Thr-phosphorylated peptides, minimizing non-specific binding and improving recovery rates. It is ideal for enriching low-abundance phosphopeptides, such as those in cell signaling pathway studies.
Fe-NTA (Iron-Nitrilotriacetic Acid): Offers low non-specific binding, making it well-suited for complex sample analysis.
MOAC (Metal Oxide Affinity Chromatography): Compatible with different phosphorylation patterns (Ser/Thr/Tyr), often used in combination with other enrichment methods.
Overall, IMAC is the most commonly used method, TiO₂ offers superior specificity, while Fe-NTA and MOAC perform better in complex sample studies. In practical applications, combining multiple enrichment strategies can enhance phosphoproteome coverage and detection sensitivity.
Q. How can the accuracy of phosphoprotein quantification be improved?
The accuracy of phosphoprotein quantification depends on experimental design, data normalization, and quantification strategies. Optimization approaches include:
1. Selecting the appropriate quantification method:
Label-Free DIA is suitable for large-scale sample analysis, combined with TIC normalization to reduce batch effects.
TMT/iTRAQ is ideal for multi-group sample comparisons but requires strict batch effect control.
2. Optimizing data normalization strategies:
TIC (Total Ion Current) normalization or iRT (indexed Retention Time) internal standards to ensure comparability across experiments.
VSN (Variance Stabilization Normalization) or Median Centering to reduce technical variation and enhance quantification stability.
3. Targeted validation:
Use PRM (Parallel Reaction Monitoring) or SRM (Selected Reaction Monitoring) to verify key phosphorylated proteins, improving data reliability.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study utilized phosphoproteomics analysis to systematically investigate the dynamic phosphorylation changes throughout the lifecycle of Saccharomyces cerevisiae, with a particular focus on the phosphorylation-mediated regulation of the fatty acid synthase (FAS) complex. Researchers identified and quantified phosphorylated proteins at different yeast growth stages, revealing that specific phosphorylation sites on the FAS complex regulate fatty acid chain length. Further experiments confirmed that these phosphorylation modifications influence the catalytic activity of FAS, thereby modulating fatty acid synthesis patterns across different growth phases. This discovery highlights the critical role of phosphorylation in lipid metabolism regulation and provides essential insights for further exploration of metabolic control, energy homeostasis, and fatty acid biosynthesis mechanisms.
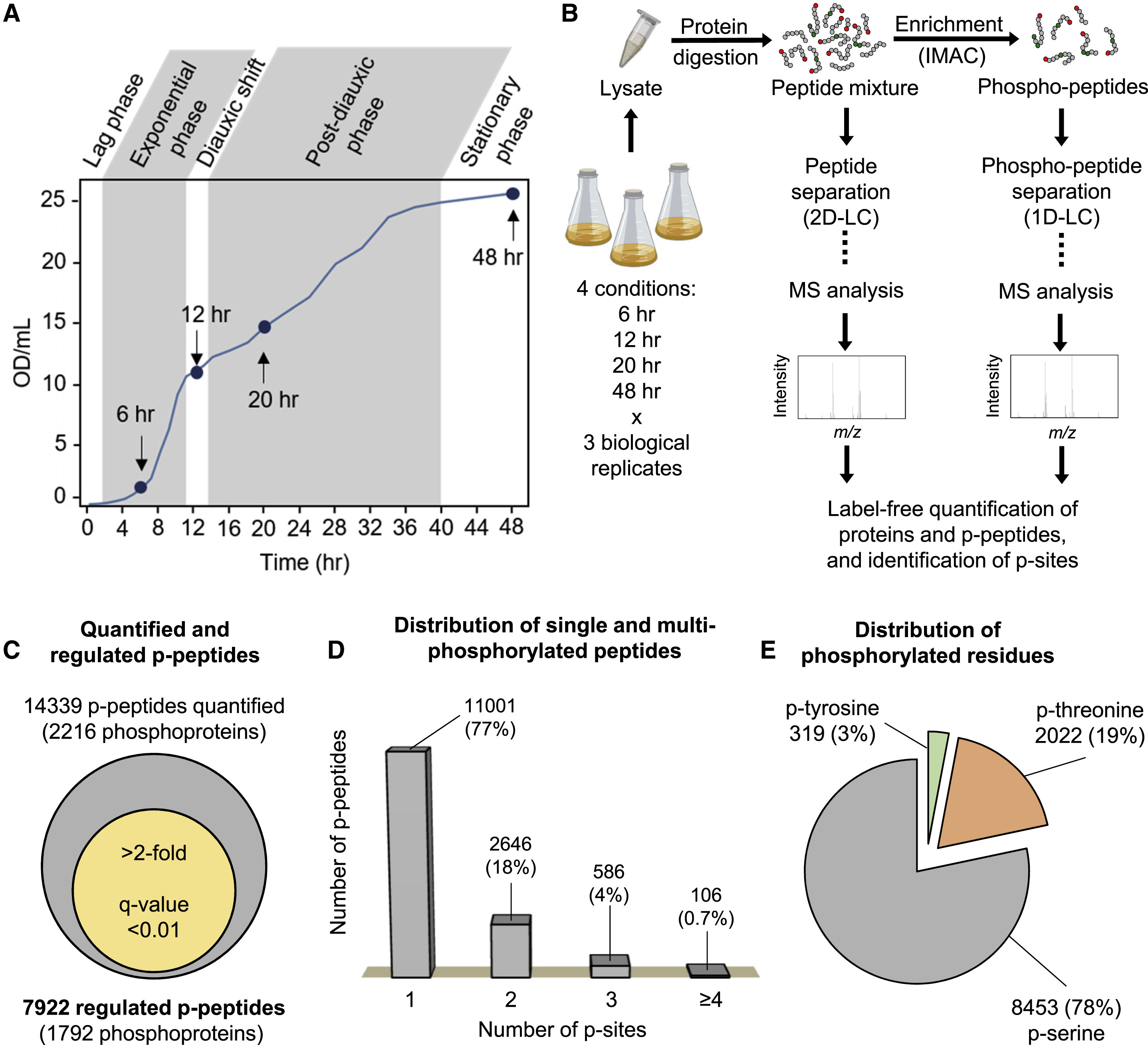
Fernando. et al. Cell Reports. 2020.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







