Peptide Drug Property Prediction Service
- Structure and sequence analysis
- Activity and ADMET prediction
- Protein–peptide interaction modeling
- Molecular dynamics simulations for conformational stability
- Drug-likeness and bioavailability scoring
- Stability testing in simulated gastric and intestinal fluids, plasma, and liver microsomes
- Permeability assays using Caco-2 and MDCK cell models
- Plasma protein binding assays
- Metabolic stability assays using human or animal hepatocytes
- Plasma concentration–time profiling
- Half-life determination in rodent models
- Organ distribution assessment using LC–MS-based quantification
- Evaluation of clearance, exposure, and accumulation patterns
MtoZ Biolabs offers a dedicated Peptide Drug Property Prediction Service that integrates in vivo, in vitro evaluation strategies, advanced computational modeling and machine-learning techniques to forecast critical physicochemical, pharmacokinetic, and pharmacodynamic features of therapeutic peptides. This service enables researchers to evaluate peptide candidates for bioactivity, ADMET profiles, and interaction potential in the early stages of drug discovery.
Background
Peptide drugs are short chains of amino acids that mimic or modulate biological signals, receptors, and enzymes. They offer unique advantages: high specificity toward targets, lower toxicity compared with small-molecule drugs, and the ability to engage challenging biological interfaces. However, peptide drugs also face distinct hurdles. They are susceptible to enzymatic degradation, often exhibit low bioavailability, and require optimization of structure, stability, and delivery.
Peptide Drug Property Prediction provides valuable insights into molecular performance by forecasting parameters such as binding affinity, solubility, permeability, stability, and bioavailability. Combining computational models with laboratory verification allows researchers to streamline candidate selection and improve success rates in early drug development.
Principle of Peptide Drug Property Prediction
MtoZ Biolabs integrates computational modeling, cell-based assays, and animal studies to deliver a full-spectrum evaluation of peptide drug properties. Our approach combines molecular simulation with laboratory validation to ensure reliability and translational value.
1. In silico (Computational) Evaluation
We apply data-driven computational models to predict molecular properties before synthesis, reducing time and cost.
2. In vitro (Experimental) Validation
Predictions are experimentally confirmed using optimized in vitro systems.
3. In vivo (Pharmacokinetic) Studies
For translational validation, MtoZ Biolabs conducts in vivo pharmacokinetic and biodistribution studies.
Peptide Drug Property Prediction Service at MtoZ Biolabs
Our core services include:
💠Peptide Drug Activity Prediction Service: Estimating the likelihood of desired biological effect based on structural and sequence features.
💠Peptide Drug ADMET Prediction Service: Forecasting absorption, distribution, metabolism, excretion, and toxicity properties to assess drug-like potential.
💠Protein-peptide Interaction Prediction Service: Predicting peptide binding to target proteins and examining potential off-target interactions.
Why Choose MtoZ Biolabs
☑️Data-Driven Reliability: Models are built using curated datasets and validated for predictive robustness.
☑️Customizable and Scalable: Service parameters and depth can be tailored to project-specific goals and peptide classes.
☑️Speed and Efficiency: Enables rapid in-silico evaluation of large peptide sets, reducing time and cost compared to purely experimental screening.
☑️Actionable Insights and Consultation: Delivered predictions include interpretive guidance and design recommendations to support informed decision-making.
☑️End-to-End Support: From initial data submission to final report delivery, MtoZ Biolabs provides clear communication and expert support.
FAQ
Q1: What types of samples are suitable?
Our service accepts peptide samples in both synthesized and recombinant forms. Clients may also submit peptide sequence data for in silico prediction without physical samples. For integrated studies, plasma or tissue-derived peptide extracts can be analyzed upon consultation.
Q2: How should I prepare my samples?
Samples should be dissolved in appropriate buffers free of salts, detergents, or stabilizing agents. Store short-term at −20°C or long-term at −80°C. For computational analysis, peptide sequences should be provided in FASTA or text format, including any known modifications.
For more information, please refer to Sample Submission Guidelines for Proteomics.
Q3: What is the service general workflow?
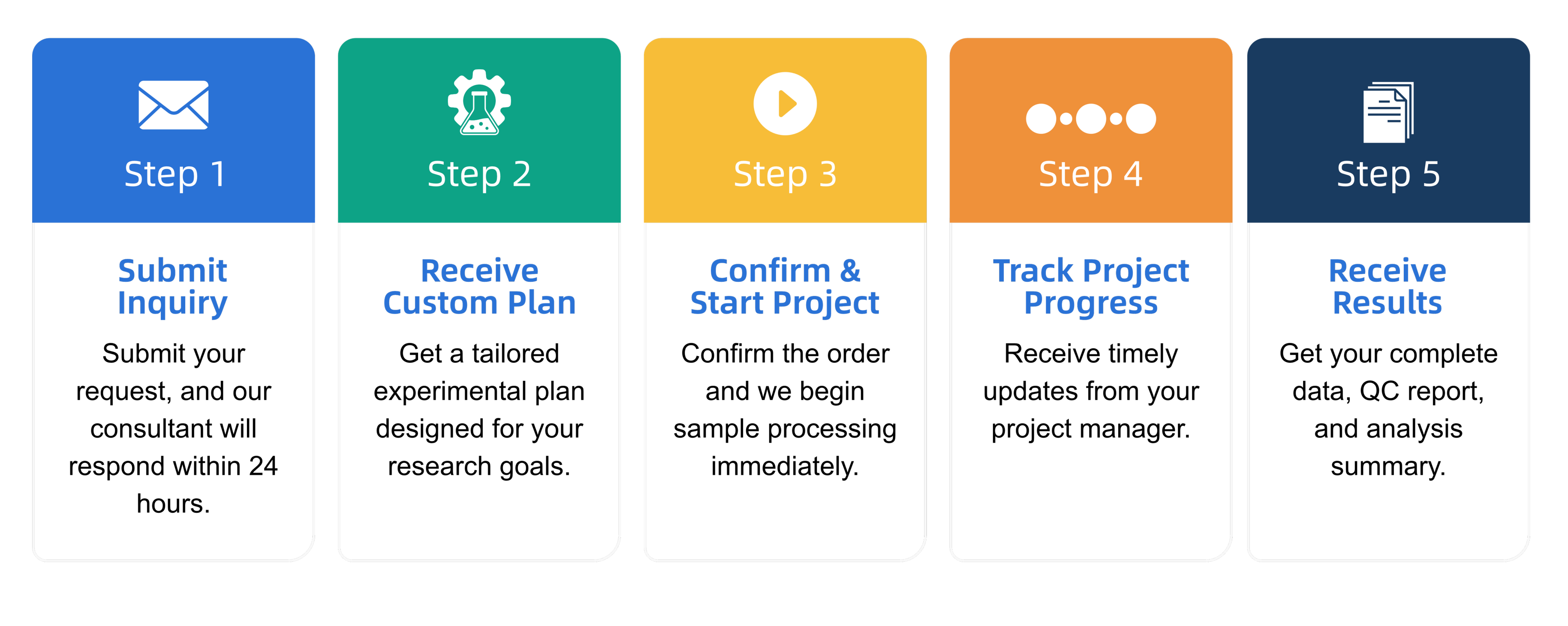
Q4: What data formats are provided?
Results are delivered in multiple formats, including:
1. Comprehensive reports in PDF format summarizing analysis and interpretations
2. Raw and processed data in Excel (XLSX) or CSV files for further analysis
3. Sequence and structural information in FASTA and PDB formats
4. Visual outputs such as correlation plots and interaction models in PNG or TIFF
5. Additional formats can be provided upon request to meet specific analysis or publication requirements.
Start Your Project with MtoZ Biolabs
Contact us today to discuss your project or request a customized proposal tailored to your research needs. Our team is dedicated to delivering accurate, data-driven insights that accelerate peptide drug discovery and optimization.
How to order?







