Oxidation Proteomics Service
- Protein Carbonylation
- Disulfide Bond Formation
- S-glutathionylation
- Methionine Oxidation
- Dityrosine Crosslinking
- Tyrosine Nitration and S-nitrosylation
- Protein Radical Modifications
Oxidation proteomics is a specialized analytical platform designed for the comprehensive identification and quantification of oxidative post-translational modifications (oxPTMs). By integrating high-resolution mass spectrometry (HRMS) with optimized derivatization-based enrichment strategies, this service enables site-specific mapping of oxPTMs and uncovers their functional roles in stress responses, disease pathogenesis, antibody stability, and food quality control.
Protein oxidation is a chemically complex process driven by reactive oxygen species (ROS) and reactive nitrogen species (RNS), primarily targeting amino acid side chains. These reactions lead to a range of covalent modifications, including carbonylation, disulfide bond formation, dityrosine crosslinking, methionine oxidation, S-glutathionylation, and nitration/nitrosylation. Such modifications alter protein structure, solubility, and activity, and have been implicated in aging, neurodegenerative diseases (e.g., Alzheimer’s, Parkinson’s), cardiovascular disorders, and muscular dystrophies. Oxidative damage also affects biopharmaceutical development (e.g., monoclonal antibodies) and food processing, compromising product quality and functional performance.
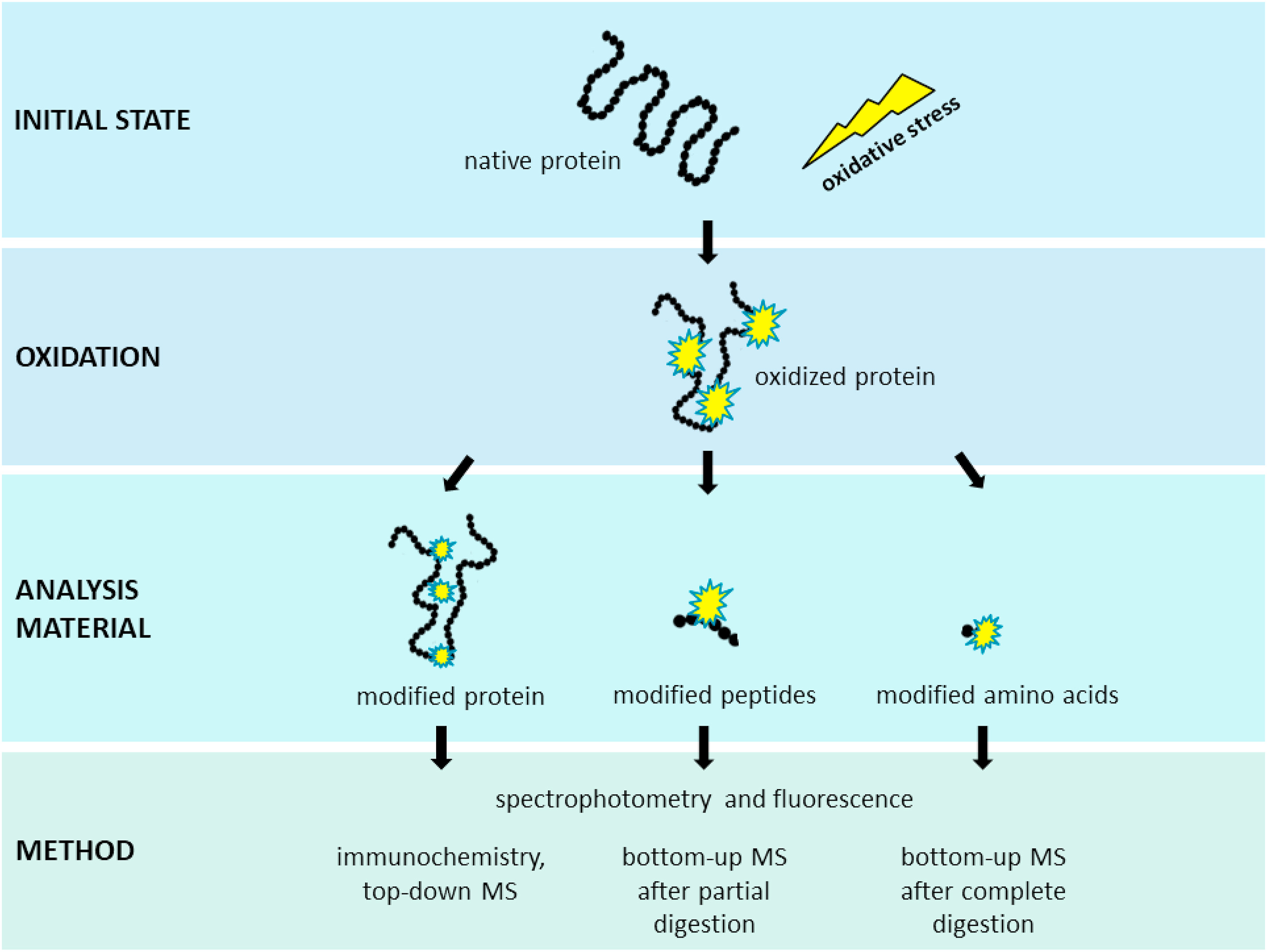
Kehm, R. et al. Redox Biology. 2021.
Figure 1. Analysis of Qxidative Protein Modifications
Powered by a state-of-the-art Nano-LC-Orbitrap mass spectrometry platform and a team with extensive expertise in PTM analysis, MtoZ Biolabs offers Oxidation Proteomics Service for detailed site identification, differential quantification, and biological interpretation of oxidative modifications across diverse research and industrial applications.
Services at MtoZ Biolabs
Utilizing high-resolution MS and robust sample preparation protocols, MtoZ Biolabs delivers precise and reproducible profiling of various oxidative modifications. Our Oxidation Proteomics Service supports the identification and quantification of the following major oxPTMs:
Additional features include quantitative analysis of modified sites, validation through spectral evidence, and in-depth functional annotation. These services are well-suited for applications in biomedical research, biotherapeutic development, and food protein quality control.
Analysis Workflow
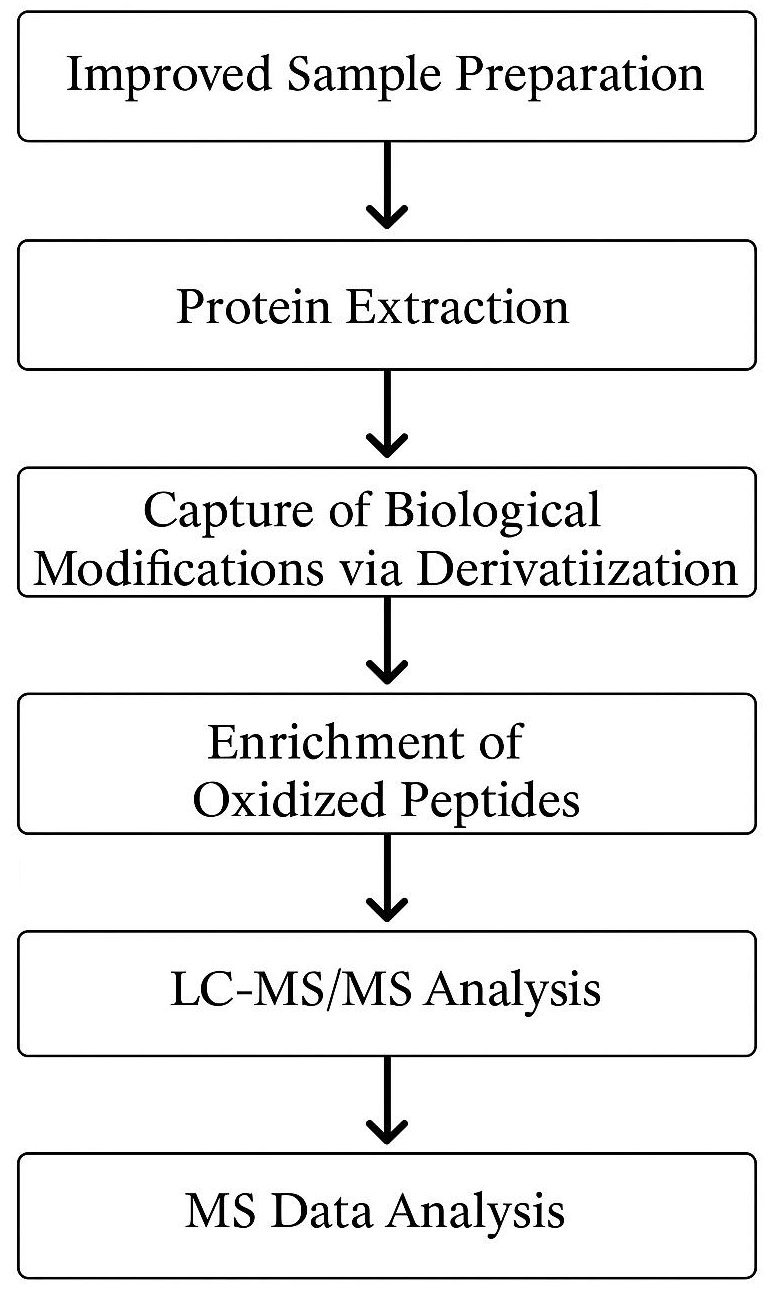
Service Advantages
✅ Advanced Mass Spectrometry Platforms
Supported by Q Exactive HF, Orbitrap Fusion, and Fusion Lumos systems to ensure sensitive detection of low-abundance oxidation sites.
✅ Comprehensive Modification Coverage
Compatible with a wide range of oxidation types, including carbonylation, disulfide bonds, S-glutathionylation, and nitration.
✅ Integrated Enrichment and Derivatization
Combined strategies enhance detection specificity and sensitivity while minimizing signal suppression and background noise.
✅ Expert Analytical Team
Proven capability in handling complex matrices such as aged tissues, pharmaceutical formulations, and processed food proteins.
✅ One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
Applications
MtoZ Biolabs' Oxidation Proteomics Service supports a wide array of research and development needs involving oxidative stress and redox biology:
· Mechanistic studies of age-related and oxidative stress-induced diseases (e.g., AD, PD, atherosclerosis, myopathies)
· Identification and validation of oxidative biomarkers for disease progression or aging
· Monitoring of oxidation sites during monoclonal antibody development and CMC workflows
· Evaluation of oxidative stability in food proteins and process optimization
· Redox profiling under drug treatment, environmental exposure, or stress challenge models
Case Study
Case 1: TMT-Based Mass Spectrometry Analysis of Protein Oxidation and Translational Regulation in Rice
Using an iodo-TMT labeling strategy, researchers systematically profiled cysteine oxidation in rice leaves following infection by Magnaporthe oryzae. The study identified 3,362 oxidized cysteine sites across 2,275 proteins involved in transcriptional regulation, ROS metabolism, and translation. Notably, 512 sites in 438 proteins exhibited elevated oxidation levels post-infection, particularly within ribosomal and transcription factor proteins.
Integration with Ribo-seq data revealed that oxidation of ribosomal proteins enhanced translation of immune-related mRNAs, contributing to host defense activation. This study highlights the role of oxidative PTMs in translational control during immune responses and demonstrates the utility of oxidation proteomics in complex biological contexts.
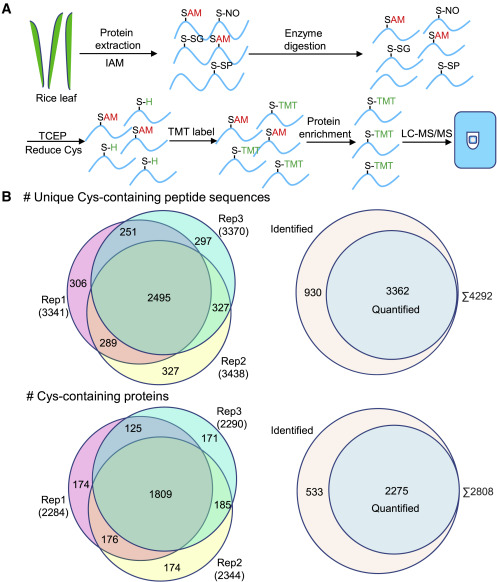
Chen, X. et al. Plant Commun. 2023.
MtoZ Biolabs leverages high-resolution MS and isobaric labeling tools (e.g., TMT) to enable precise site-level quantification and biological interpretation of oxidation events, empowering deeper investigation into redox signaling in health and disease.
FAQ
Q1: How does MtoZ Biolabs ensure preservation of native oxidative modifications during sample processing?
We utilize protective measures such as low-temperature lysis, rapid extraction, thiol-blocking reagents (e.g., NEM or IAM), and non-reducing buffers to minimize artificial oxidation or loss of endogenous modifications, ensuring data integrity and reproducibility.
Q2: Is the service suitable for comparative studies with multiple sample groups?
Yes. We offer both label-free and isobaric labeling strategies (TMT/iTRAQ) for quantitative comparisons across multiple experimental groups. The results support pathway enrichment, oxidative modification profiling, and mechanistic modeling.
MtoZ Biolabs is committed to delivering high-quality, customizable solutions for post-translational modification analysis. Our Oxidation Proteomics Service helps researchers unravel the complexity of oxidative modifications and their biological consequences—advancing discovery in disease biology, therapeutic development, and food science. For consultation or collaboration, please visit our website or contact your dedicated project advisor.
How to order?







