Nitration Proteomics Service
Nitration proteomics analysis is a high-resolution proteomic detection and analysis technology focusing on protein nitration modification. Protein nitration primarily refers to the formation of 3-nitrotyrosine on tyrosine residues under the action of reactive nitrogen species (RNS). This process is typically mediated by strong oxidants such as peroxynitrite (ONOO⁻), generated from the reaction between nitric oxide (NO) and superoxide anion (O₂⁻). Such modifications can alter protein conformation, activity, interactions, and stability, thereby regulating cellular signaling pathways, metabolic processes, and immune functions. This service integrates multiple analytical strategies with bioinformatics to enable accurate identification and quantification of nitration sites.
Nitration proteomics service is widely applied in studies of cardiovascular diseases, neurodegenerative disorders, cancer, inflammatory responses, and environmental toxicology. It can be used to explore protein nitration profiles under pathological conditions, identify potential biomarkers or therapeutic targets, and support validation of drug mechanisms and intervention strategies. This service provides a powerful tool for elucidating the regulatory role of nitration in disease-related processes.
Services at MtoZ Biolabs
Based on advanced analytical platforms and comprehensive enrichment strategies, MtoZ Biolabs offers the nitration proteomics service focuses on the detection and quantification of nitrification modifications in proteins. This service enables high-sensitivity identification of nitration sites in cells, tissues, and purified protein samples, delivering high-quality data including modified peptide sequences, site localization, spectral information, and relative abundance. The data support studies on disease mechanisms, oxidative stress, and potential therapeutic target identification. The analytical methods include the following approaches:
1. Mass Spectrometry (MS)
Using high-resolution LC-MS/MS platforms, this method detects nitrated peptides, particularly 3-nitrotyrosine modifications on tyrosine residues. It enables precise identification of modification sites, peptide sequences, and relative abundance. Coupling with anti-3-nitrotyrosine antibody enrichment improves the detection of low-abundance peptides, making this the most sensitive and widely used approach in nitration proteomics.
2. High-Performance Liquid Chromatography (HPLC)
HPLC is commonly used in conjunction with mass spectrometry or as a sample pretreatment step to separate proteins or peptides with different modification states. It supports the identification of nitration signatures and can be applied for semi-quantitative analysis.
3. Nuclear Magnetic Resonance (NMR)
NMR is suitable for structural analysis of purified proteins. It can directly observe chemical environment changes caused by nitration, allowing precise localization of modified residues. However, due to its relatively low sensitivity, it is typically used for validating nitration on specific proteins.
4. Immunodetection Methods (Western Blot/ELISA)
These methods employ anti-3-nitrotyrosine antibodies to detect nitrated proteins. They are suitable for preliminary screening or validation of known nitrated targets. While easy to perform, they do not provide site-specific modification information.
Analysis Workflow
1. Protein Extraction and Digestion
Proteins are extracted from cells, tissues, or purified samples, followed by reduction and alkylation. The proteins are then digested with trypsin to generate peptides suitable for subsequent analysis while preserving nitration modifications.
2. Modified Peptide Enrichment
Nitrated peptides are selectively captured using strategies such as anti-3-nitrotyrosine antibody enrichment, chemical derivatization labeling, or solid-phase extraction, effectively enhancing the detection of low-abundance modifications.
3. Multiplex Platform Detection
According to research needs, a combination of LC-MS/MS, HPLC, NMR, and other analytical techniques is applied to accurately identify nitration sites, obtain peptide sequences, characterize modifications, quantify abundance changes, and acquire structural information, ensuring comprehensive and accurate data.
4. Data Processing and Result Output
Dedicated databases and algorithm tools are used to identify and quantify modification sites. A complete report is provided, including spectra, modification positions, relative abundance, and functional annotations, supporting mechanism research and biomarker discovery.
Sample Submission Suggestions
1. Sample Type
A wide range of sample types is supported, including cell lysates, tissue homogenates, serum/plasma, and purified proteins.
2. Buffer System Requirements
Avoid using buffers containing high salt, SDS, glycerol, EDTA, Tris, or other components that may interfere with mass spectrometry detection or antibody-based enrichment.
3. Sample Storage and Transportation
Samples should be stored at –80°C and protected from repeated freeze-thaw cycles to prevent loss of modifications or protein degradation. During transportation, dry ice must be used to ensure the stability of the modification state.
Service Advantages
1. High-Sensitivity Detection Platform
Powered by high-resolution mass spectrometry systems such as Orbitrap, combined with advanced chromatographic and enrichment technologies, this platform enables accurate identification of low-abundance nitration sites, suitable for complex sample analysis.
2. Multi-Strategy Detection Approaches
Integrates multiple analytical techniques including mass spectrometry, immunoaffinity enrichment, HPLC, and NMR to achieve comprehensive qualitative and quantitative analysis of nitrated proteins, enhancing data accuracy and completeness.
3. Dedicated Enrichment Methods
Utilizes anti-3-nitrotyrosine antibodies or specific chemical derivatization strategies to enrich target modified peptides, effectively improving detection rates and supporting various sample types and research scenarios.
4. End-to-End Technical Support
Covers the full workflow from sample preparation, peptide enrichment, multi-platform detection to data analysis and biological annotation, helping researchers systematically investigate the functional roles of nitration in disease and signaling regulation.
Applications
1. Oxidative Stress-Related Disease Research
Nitration is a critical marker of oxidative stress and is widely present in pathological processes such as cardiovascular diseases and neurodegenerative disorders (e.g., Parkinson's disease, Alzheimer's disease). It is suitable for investigating the mechanistic roles of nitration in disease onset and progression.
2. Inflammation and Immune Regulation Research
The nitration proteomics service can be used to explore the role of 3-nitrotyrosine in immune cell activation, inflammatory signaling regulation, and imbalance of inflammatory responses, aiding in the identification of molecular features associated with immune dysfunction.
3. Drug Mechanism and Target Validation
By comparing nitration levels across differently treated sample groups, the service allows evaluation of how drugs affect protein nitration status, supporting target confirmation and mechanism-based studies.
4. Environmental Toxicology and Pollutant Response Research
The nitration proteomics service enables the assessment of nitration responses induced by pollutants (such as NO₂ and peroxynitrite) and their impact on cellular functions, revealing protein modification changes and toxicity mechanisms under environmental exposure.
Case Study
1. Proteomics Quantification of Protein Nitration
This study aims to establish a high-throughput, sensitive, and quantitative method for analyzing protein nitration modifications, specifically to systematically characterize the distribution and variation of 3-nitrotyrosine modifications. Human cell samples subjected to oxidative stress were used as the study model. Anti-nitrotyrosine antibody enrichment, tandem mass tag (TMT) labeling, and high-resolution LC-MS/MS were employed to identify and quantify nitrated peptides. As a result, hundreds of high-confidence nitration sites were identified, revealing their dynamic regulatory roles in multiple stress-related signaling pathways. The findings demonstrate that this method possesses excellent throughput, sensitivity, and reproducibility, and can be widely applied to the study of nitration modifications and the discovery of potential therapeutic targets in pathological conditions such as inflammation, cardiovascular diseases, and neurodegenerative disorders.
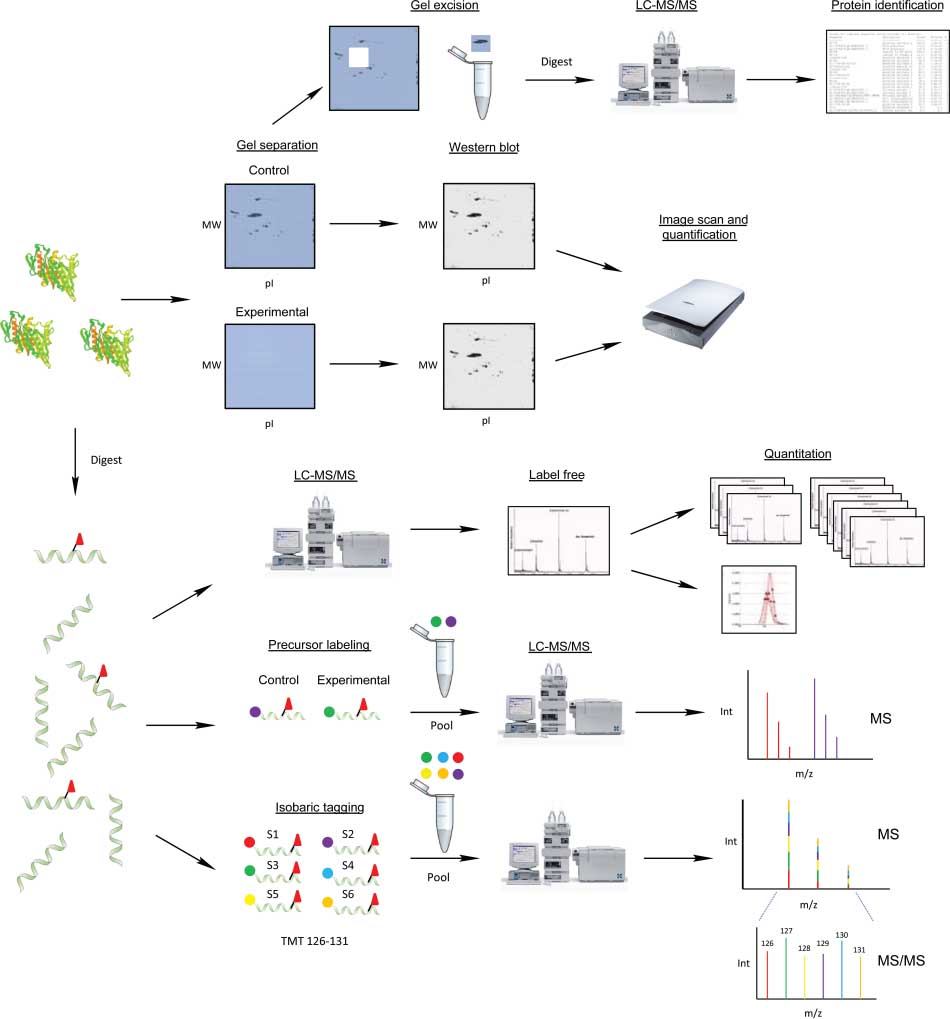
Evan, A R. et al. Reviews in Analytical Chemistry, 2013.
Figure 1. Schematic of General Workflows for Quantitative Proteomic Studies of Nitrated Proteins.
2. Proteomic Identification of Nitrated Proteins in Alzheimer's Disease Brain
This study aims to identify protein nitration modifications in the brain tissues of Alzheimer's disease (AD) patients using proteomic approaches, in order to explore their potential roles in disease pathogenesis. Cortical samples from AD patients and control individuals were analyzed. Nitrated proteins were enriched by immunoprecipitation, followed by identification using SDS-PAGE and mass spectrometry. The results revealed significantly elevated levels of protein nitration in AD brain tissues, with numerous specific nitration sites observed in proteins related to energy metabolism, cytoskeletal structure, and antioxidant systems. The study concludes that protein nitration may contribute to the pathological progression of AD by altering the structure and activity of key functional proteins, highlighting its important role in neurodegenerative diseases and its potential as a diagnostic or therapeutic target.
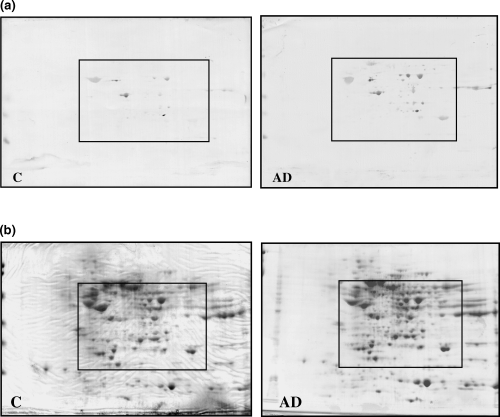
Castegna, A. et al. Journal of Neurochemistry, 2003.
Figure 2. Two-Dimensional 3NT Blots (a) and Comassie Blue-Stained Gels (b) from Control and AD Brain Extracts.
FAQ
Q1: Does the Service Provide Quantitative Analysis?
A1: Yes, we support multiple quantitative approaches including Label-Free and TMT, enabling relative comparison of nitration levels between different sample groups. This is suitable for studies involving dynamic regulation or drug intervention analysis.
Q2: Does the Service Include Bioinformatics Analysis Results?
A2: Yes. We provide a comprehensive report that includes a list of modification sites, quantitative spectra, functional annotations (such as GO and KEGG), and pathway enrichment analysis of modified proteins.
How to order?







