Molecular Docking Service
- Structure files (PDB)
- Docking conformations and energy scoring results (TXT or CSV)
- Interaction analysis and visualization figures (PNG or PDF)
- Comprehensive analysis report (PDF)
- Raw computational files (compressed format)
MtoZ Biolabs employs an advanced computational simulation platform and has launched the molecular docking service which enables systematic analysis of binding modes, interaction features, and potential key sites between active peptides and various receptor molecules. This service is widely applied in active peptide screening, structural optimization, and mechanism studies, providing a reliable data foundation for functional peptide development, targeted design, and activity prediction.
What Is Molecular Docking?
Molecular docking is a computational technique used to predict the optimal binding modes between molecules and to analyze their interaction relationships. Its principle is to simulate possible ligand binding conformations on a receptor through spatial matching and energy scoring, and to evaluate binding stability and affinity. As an important tool in structural biology and drug design, molecular docking can rapidly reveal potential interaction patterns between peptides and proteins, receptors, or small molecules. For active peptide research, it helps identify key binding sites, predict functional mechanisms, and support functional peptide screening and peptide drug sequence optimization.
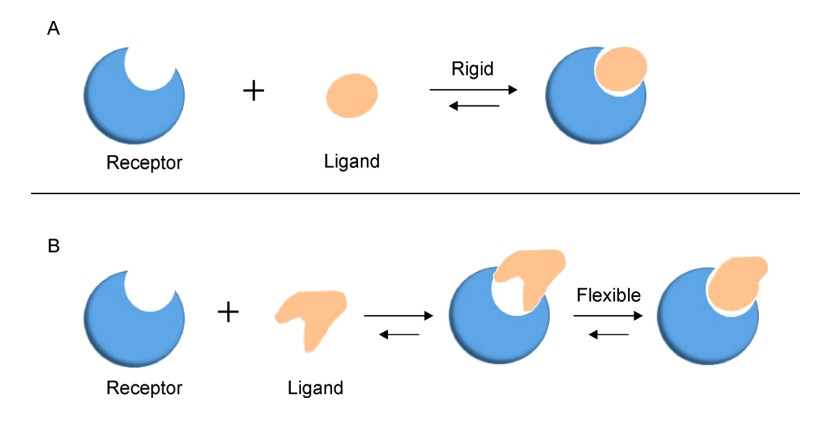
Fan, J Y. et al. Quantitative Biology, 2019.
Figure 1. Two Models of Molecular Docking.
Molecular Docking Service at MtoZ Biolabs
1. Peptide-Protein Receptor Docking Analysis
Simulates the binding mode between active peptides and protein receptors, identifies key interaction sites, and evaluates binding stability.
2. Peptide-Small Molecule Docking Study
Predicts the binding conformations and energy scores between peptides and small molecules, supporting the exploration of potential regulatory or competitive mechanisms.
3. Peptide-Drug Molecule Docking Evaluation
Analyzes the interaction characteristics between peptides and candidate drug molecules, assisting in studying pharmacological mechanisms and optimizing peptide-based therapeutics.
4. Peptide-Nucleic Acid Docking Analysis
Simulates the binding interactions between peptides and DNA or RNA, revealing the structural basis and functional patterns of nucleic acid-binding peptides.
5. Peptide-Lipid or Membrane Model Docking Study
Predicts the interactions between peptides and lipid bilayers, membrane proteins, or membrane models, supporting the analysis of functional characteristics of antimicrobial peptides, transmembrane peptides, and related molecules.
6. Peptide-Ligand Pocket Binding Mode Prediction
Simulates how peptides fit into receptor binding pockets, evaluates affinity, identifies key residues, and supports functional screening.
7. Peptide-Peptide Docking Conformation Analysis
Predicts the complex structures and stability of peptide-peptide interactions, providing structural evidence for studying synergistic effects or designing composite peptides.
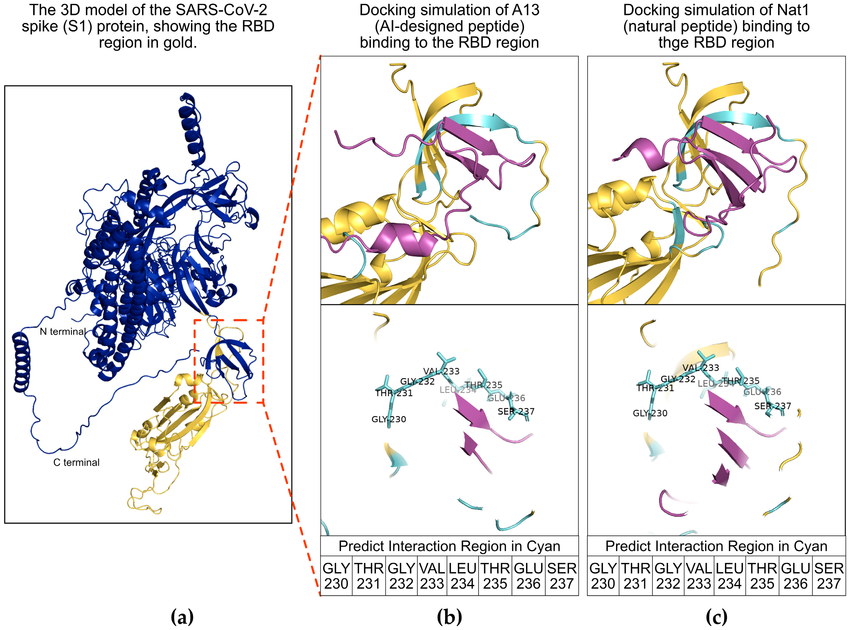
Wang, J S. et al. International Journal of Molecular Sciences, 2025.
Figure 2. Molecular Docking Analysis of Peptides with the SARS-CoV-2 S1 Protein Receptor-Binding Domain Region.
Why Choose MtoZ Biolabs?
✅ High-Accuracy Prediction: Accurately simulates the binding modes of active peptides based on reliable algorithms and optimized parameters.
✅ Rapid Energy Scoring: Provides energy calculations and ranking for multiple models, enabling efficient screening of high-affinity conformations.
✅ Flexible Customized Analysis: Adjusts docking strategies and parameters according to type, research objectives, and target molecule characteristics.
✅ Efficient Computational Workflow: A standardized computational pipeline shortens analysis time and rapidly delivers actionable results.
✅ One-Stop Analysis: Covers the entire workflow from structure preparation to result interpretation, reducing user workload.
Applications of Molecular Docking Service
1. Functional Peptide Screening and Activity Prediction
Used to evaluate the binding potential of active peptides with different receptor molecules and rapidly identify candidate sequences with biological activity.
2. Peptide-Ligand Competition and Synergy Study
Used to analyze competitive or synergistic interactions between peptides and other ligands at the same binding site, supporting combination strategy design.
3. Ligand-Receptor Specificity Analysis
Used to assess the binding differences of molecules with different structural classes (peptides, small molecules, drug molecules, etc.) toward the same target, supporting selectivity studies and target validation.
4. Pathway-Related Interaction Prediction
Predicts interactions between peptides and various regulatory proteins or small molecules based on pathway-related key nodes, aiding mechanism inference.
5. Protein Structure Mutation Effect Analysis
Used to simulate changes in peptide or ligand binding resulting from mutations in the target protein, enabling evaluation of mutation impact and potential resistance risks.
Deliverables
1. Comprehensive Experimental Details
2. Peptide and Receptor Structure Models
3. Docking Conformations and Energy Scoring Results
4. Interaction Analysis and Visualization Data
5. Comprehensive Analysis Report
6. Raw Data Files
FAQ
Q1: What types of samples are suitable?
A1: Applicable to peptides with clearly defined structural information and their docking counterparts, including proteins, small molecules, drug molecules, nucleic acids, and membrane structure models. If the receptor or peptide lacks a three-dimensional structure, sequence information can be provided and we will assist in building a predicted model.
Q2: What is the service general workflow?
A2:
Q3: What data formats are provided?
A3: The deliverables include:
If special analytical requirements exist, data formats can be customized according to project specifications.
Q4: How should I prepare the samples?
A4: To ensure the accuracy of docking analysis, it is recommended to follow the guidelines below:
(1) Providing Structure Files: If three-dimensional structures are available, submit the PDB files of the peptide and receptor; if no structures exist, sequences may be provided for model prediction.
(2) Supplementing Background Information: Including functional data, potential binding sites, or experimental observations helps optimize the docking strategy.
(3) Clarifying Research Objectives: Specify the docking target, research purpose (such as screening, optimization, or mechanism study), and key points of interest to support development of an appropriate analysis plan.
For more information, please refer to Sample Submission Guidelines for Proteomics, Sample Submission Guidelines for Metabolomics.
Start Your Project with MtoZ Biolabs
Contact us to discuss your experimental design or request a quote. Whether you are exploring the binding mode between peptides and specific molecules or studying the structural mechanisms behind molecular interactions, MtoZ Biolabs can provide reliable and systematic docking analysis support.
How to order?







