Model Reconstruction of Crystal Structures Using Cryo-EM and NMR
Model Reconstruction of Crystal Structures Using Cryo-EM and NMR is a service that integrates atomic resolution three-dimensional density maps provided by cryo-electron microscopy (Cryo-EM) with chemical shift, dipolar coupling, NOE, and other constraint information from nuclear magnetic resonance (NMR) spectroscopy. This enables reconstruction, completion, or conformational refinement of crystal structures, ultimately generating more accurate and biologically relevant structural models.
Crystal structure determination is a key technique for understanding the functional mechanisms of biological macromolecules, intermolecular interactions, and drug design. X-ray crystallography has long been considered the “gold standard” for resolving the structures of biomolecules such as proteins. However, despite its excellent performance under large crystal conditions, it faces major limitations in systems with microcrystalline samples, strong heterogeneity, or highly flexible regions.
Cryo-EM and NMR spectroscopy are complementary in terms of spatial resolution and dynamic structure characterization. Cryo-EM provides high-resolution crystal density maps, while NMR contributes information on flexible regions and solution-state conformations. By integrating structural data from both techniques, more accurate and comprehensive crystal structure model reconstruction can be achieved—particularly for systems with high structural heterogeneity, small molecule binding, large complexes, or locally disordered regions.
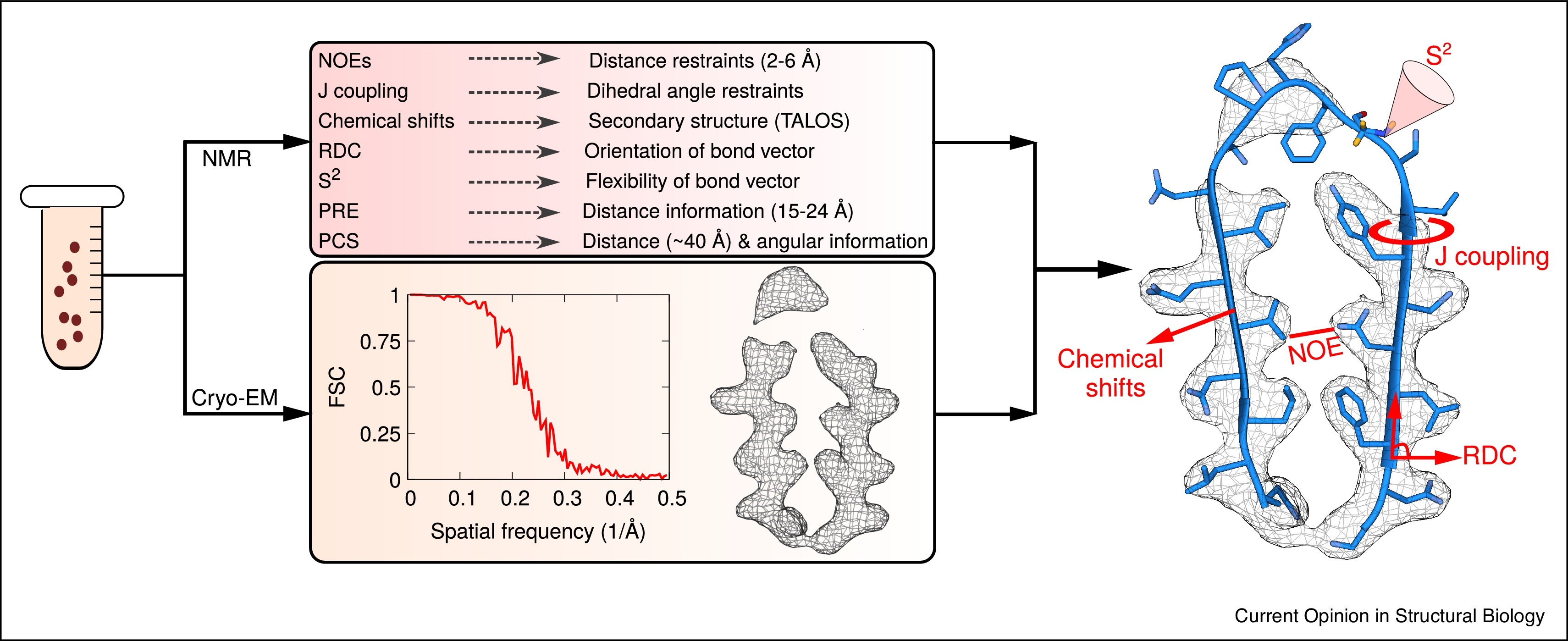
Geraets JA. et al. Curr Opin Struc Biol. 2020.
Based on advanced Cryo-EM and NMR platforms, MtoZ Biolabs provides Model Reconstruction of Crystal Structures Using Cryo-EM and NMR to accurately resolve atomic-level structural features of crystals. This service helps researchers overcome the technical limitations of traditional X-ray crystallography by systematically identifying flexible regions, ligand-bound conformations, and dynamic structural states. It is widely applicable in key research areas such as protein microcrystal modeling, natural product structure confirmation, drug polymorph analysis, and structure-based drug design.
Analysis Workflow
The core analytical workflow of Model Reconstruction of Crystal Structures Using Cryo-EM and NMR includes:
1. Sample Preparation
Prepare high-quality samples that meet the requirements for both Cryo-EM and NMR analysis.
2. Cryo-EM Imaging and 3D Reconstruction
Acquire Cryo-EM images and reconstruct high-resolution three-dimensional density maps.
3. NMR Data Acquisition and Constraint Extraction
Perform multidimensional NMR experiments to obtain spatial constraints for key structural regions.
4. Structure Integration and Model Building
Integrate Cryo-EM density and NMR constraints to complete and refine the crystal structure model.
5. Model Optimization and Result Validation
Use modeling and validation tools to optimize structure quality and ensure consistency with experimental data.
6. Structural Interpretation and Functional Analysis
Deliver atomic-level structural models, density maps, modeling workflow documentation, and insights into conformational variations and potential functional mechanisms.
Applications
Typical applications of Model Reconstruction of Crystal Structures Using Cryo-EM and NMR include:
Structural Completion and Conformational Refinement
Ideal for completing missing loops, flexible regions, or disordered fragments in crystal structures to improve structural integrity.
Dynamic Structure Analysis
Enables the identification of conformational changes under different states and reveals transition mechanisms during functional cycles of proteins or complexes.
Validation of Crystal Structure Authenticity
Combines solution-state NMR and near-native Cryo-EM data to evaluate whether crystal structures are subject to environment-induced conformational bias.
Ligand Binding and Functional Site Investigation
Assists in identifying ligand or biopolymer binding sites and assessing conformational adaptability, supporting structure-based functional interpretation and drug screening.
Cross-platform Data Integration and Multiscale Modeling
Integrates data from crystallography, Cryo-EM, and NMR to construct physiologically relevant models, especially for large complexes and membrane protein systems.
FAQ
Q. What Types of Samples are Suitable for this Service? Is Crystallization Required?
This service is mainly applicable to microcrystalline or nanocrystal samples, especially those not amenable to traditional X-ray crystallography—such as crystals smaller than 10 µm, proteins with flexible domains, or systems with high crystallization heterogeneity.
Large single crystals are not required, but the sample must have good crystallinity and stability to support Cryo-EM imaging. NMR is particularly useful for systems with disordered regions or significant conformational variability.
Q. How are Cryo-EM and NMR Data Integrated for Structure Modeling?
MtoZ Biolabs applies a hybrid modeling strategy: Cryo-EM provides the global density framework for backbone and ordered region modeling, while NMR provides NOE, RDC, J-coupling, and other conformational constraints to refine flexible loops and side chains. Through integrated modeling tools such as Phenix and Rosetta-NMR, the Cryo-EM density and NMR constraints can be coordinated and optimized to build a high-precision atomic model that fits the experimental data.
Case Study
This study used a combination of Cryo-EM and NMR to reconstruct the structure of the Clr4 methyltransferase complexed with an H3K9-methylated nucleosome. Cryo-EM first resolved the 3D conformation of Clr4 bound to the H3K9me nucleosome, showing that Clr4 recognizes methylated H3K9 via its chromodomain (CD) and stabilizes its chromatin localization through auxiliary contacts mediated by its disordered tail. NMR experiments provided additional molecular details of the interactions between the disordered region and the nucleosome surface, revealing a dynamic assembly process from the unmethylated to the primed methylated state. The final model demonstrated that Clr4 mediates heterochromatin establishment through a “bridging recognition–initial modification” mechanism, offering key structural insights into the selective recognition mechanism of histone-modifying enzymes and the epigenetic regulatory network.
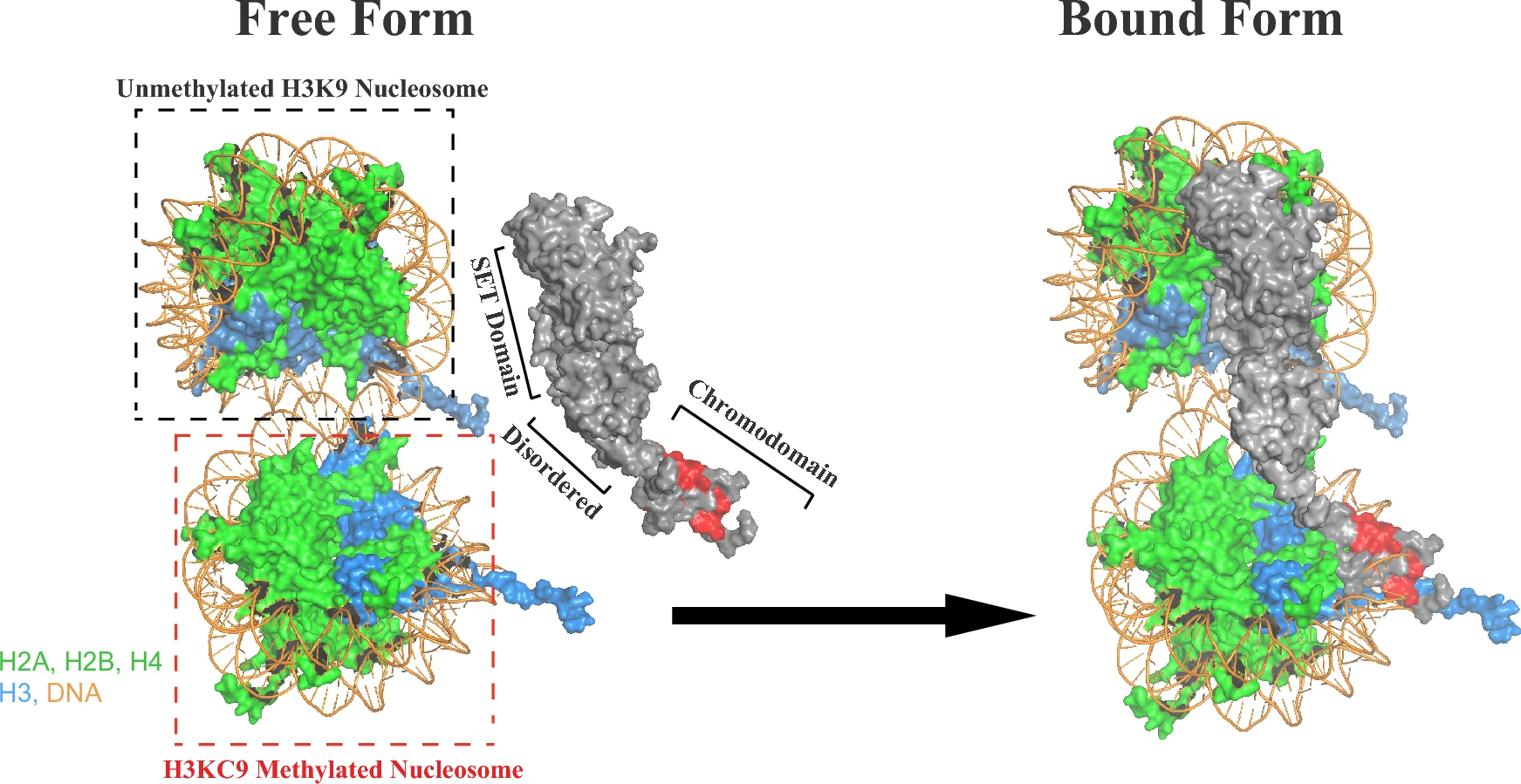
Saab C. et al. Sci Rep. 2024.
How to order?







