microRNA Sequencing Service
- Cells: ≥ 1 × 10⁷
- Fresh plant tissue: ≥ 500 mg
- Animal tissue: ≥ 300 mg
- Blood: ≥ 2 mL
- RNA Samples: Total RNA ≥ 3 μg
- Store at –80 °C and ship on dry ice
MicroRNA (miRNA) sequencing is a next-generation sequencing (NGS) technology designed for the comprehensive analysis of small RNA populations, particularly miRNAs. miRNAs are endogenously encoded, non-coding, single-stranded RNA molecules that are typically 20 to 25 nucleotides in length. miRNAs are highly conserved across species and play a central role in post-transcriptional gene regulation by binding to target messenger RNAs (mRNAs) to either inhibit their translation or promote their degradation. With the advancement of high-throughput sequencing platforms, miRNA sequencing allows researchers to directly profile all miRNAs within a specific size range in biological samples—without the need for prior sequence information. This makes it possible to not only quantify known miRNAs but also to identify and characterize novel miRNA species, supporting diverse applications in gene expression regulation, developmental biology, disease biomarker discovery, and therapeutic target research.
Service at MtoZ Biolabs
MtoZ Biolabs offers comprehensive microRNA Sequencing Services designed to support researchers in decoding small RNA expression landscapes across diverse biological systems. By integrating optimized small RNA library preparation protocols with advanced high-throughput sequencing platforms, we enable accurate sequencing of miRNAs and facilitate the discovery and characterization of novel miRNA species. Our end-to-end solution covers everything from total RNA quality assessment and small RNA enrichment to sequencing and downstream bioinformatics analysis. Whether you're studying disease-associated miRNA signatures, regulatory networks, or post-transcriptional gene silencing mechanisms, MtoZ Biolabs provides accurate, high-resolution data tailored to your experimental goals.
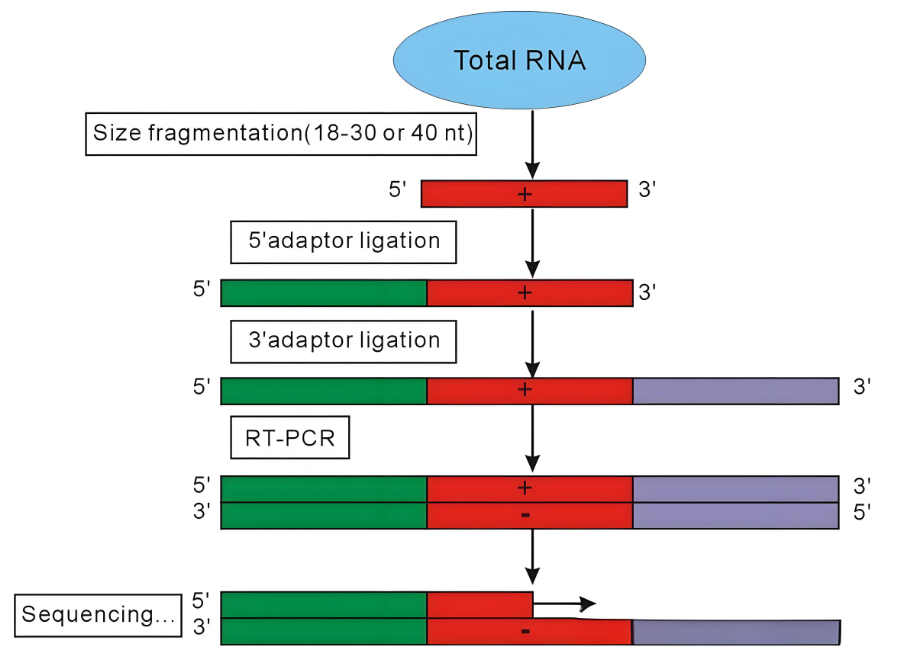
Figure 1. Flowchart Depicting miRNA Sample Preparation and Sequencing
Technical Principles
miRNA sequencing is a molecular biology technique based on high-throughput sequencing platforms, designed to deeply analyze intracellular miRNAs. The core principle involves isolating small RNA molecules, typically 20–24 nucleotides in length, from total RNA using either sequence-specific oligonucleotide probes or miRNA-enrichment strategies, followed by sequencing to identify and characterize miRNA profiles.
1. Sequence-Specific Oligonucleotide Probe-Based Capture
This approach uses short DNA or RNA probes that are designed to be fully complementary to specific miRNA sequences. These probes are immobilized on solid supports such as microarray chips or magnetic beads. When total RNA is incubated with these immobilized probes, target miRNAs hybridize to their complementary sequences due to base-pairing specificity, while non-target RNAs are washed away. This enables high-specificity enrichment of miRNAs prior to sequencing.
2. miRNA-Specific Enrichment Techniques
Several biochemical methods are employed to selectively capture miRNAs:
💠Poly(A) Tailing: Since native miRNAs lack a poly(A) tail, an artificial poly(A) sequence can be enzymatically added. This tail allows miRNAs to be captured using oligo(dT) magnetic beads that bind the added poly(A) sequence.
💠Molecular Tagging: Using enzymes such as T4 RNA ligase, a molecular tag (e.g., biotin) can be attached to either the 3' or 5' end of miRNAs. These tagged molecules can then be isolated using streptavidin-coated magnetic beads that selectively bind to the biotin label.
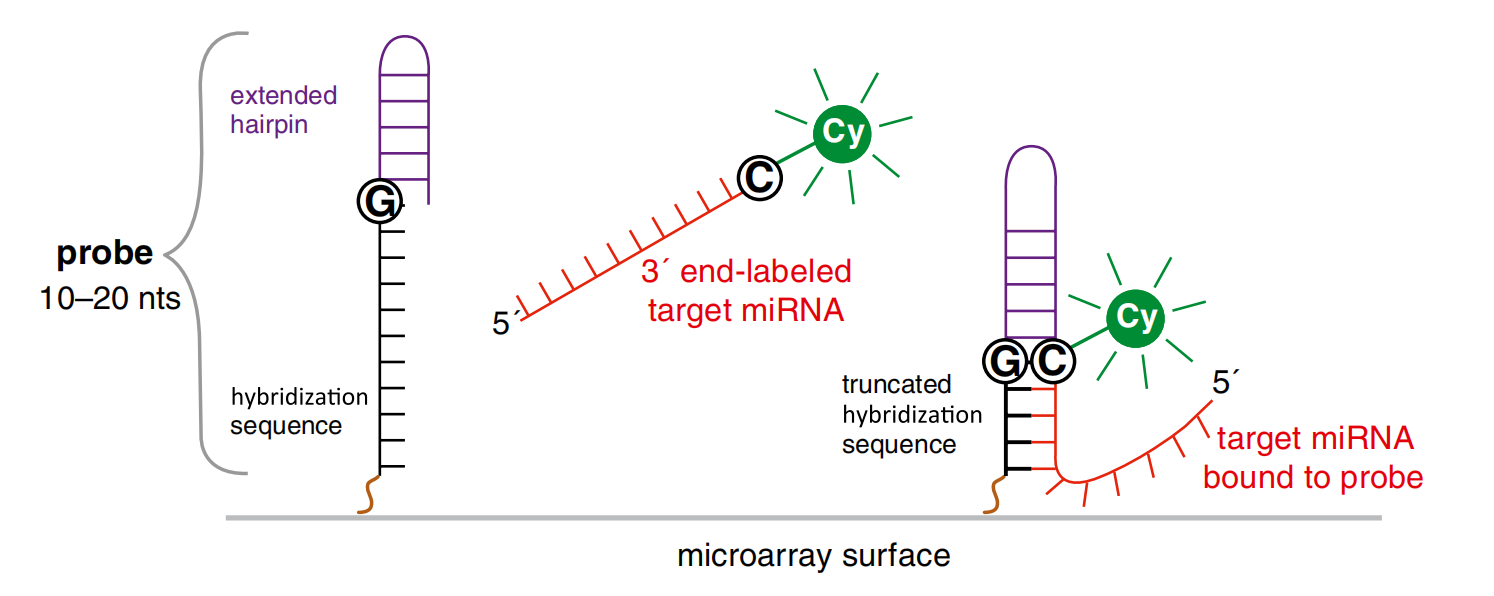
White, P. 2011.
Figure 1. miRNA Microarray Probe Design
Service Advantages
☑️High Specificity: Custom-designed primers and probes enable precise identification of target miRNA sequences, minimizing background noise.
☑️High Throughput: Allows simultaneous detection of tens of thousands of miRNAs, providing comprehensive profiling of complex miRNA expression patterns.
☑️High Sensitivity: Capable of detecting low-abundance miRNAs with high accuracy, even in complex biological samples.
☑️Broad Dynamic Range: Effectively captures miRNAs with a wide range of expression levels, from low to highly expressed targets.
☑️Sample Versatility: Compatible with a wide variety of biological sample types, including blood, tissues, cell extracts, and body fluids.
☑️Data Comparability: Sequencing results are standardized and reproducible across experiments and laboratories, facilitating data integration and sharing.
☑️In-Depth Analysis: Enables advanced bioinformatics analysis to uncover the relationships between miRNAs and disease mechanisms, cellular functions, and biological pathways.
Applications
1. Medical Diagnostics
Enables early detection and classification of diseases such as cancer, cardiovascular disorders, and neurological conditions through miRNA biomarker profiling.
2. Drug Discovery and Development
Assists in identifying therapeutic targets, evaluating drug efficacy, and predicting off-target effects via miRNA expression analysis.
3. Functional Genomics
Facilitates the study of gene regulatory networks and post-transcriptional mechanisms in diverse biological systems.
4. Agricultural Biotechnology
Supports crop improvement, disease resistance research, and trait selection through miRNA expression profiling in plants.
5. Environmental and Ecotoxicology
Aids in monitoring environmental changes and evaluating toxicological impacts using miRNA as sensitive molecular indicators.
Sample Submission Suggestions
1. Sample Types
We accept various sample types, including but not limited to:
2. Storage & Shipping
*Note: Please contact us before shipping for any special requirements or pre-analysis consultation.
Case Study
Prostate cancer (PCa) is one of the most common malignancies among men and the second leading cause of cancer-related death in males. Advanced metastatic PCa frequently spreads to bone, with over 90% of patients showing skeletal metastases upon autopsy. These lesions often display osteolytic characteristics, and pathological fractures are common, indicating a disruption of bone homeostasis. In this study, researchers unexpectedly identified prostate cancer-derived exosomes as key modulators of bone remodeling, capable of inducing osteolytic lesions and altering the bone microenvironment. Exosomes isolated from osteoblastic, osteolytic, and mixed-type PCa cell lines were shown to promote osteoclast differentiation in vitro and drive bone resorption in vivo.
Mechanistically, miR-92a-1-5p, the most abundant miRNA among those transported by prostate cancer-derived exosomes, played a central role by directly targeting COL1A1, resulting in the downregulation of type I collagen expression. This reduction not only promoted osteoclast differentiation but also inhibited osteoblast activity, ultimately tipping the balance toward bone resorption. In addition, prostate cancer-derived exosomes significantly reduced the expression of type I collagen in vivo. This study highlights the critical role of tumor-derived exosomal miRNAs in modulating bone homeostasis and suggests new therapeutic targets for managing skeletal metastasis in prostate cancer.
Through the integration of cell culture, exosome isolation and characterization, exosomal uptake assays, differentiation studies, and murine models, the study provides compelling evidence for the regulatory role of miRNAs in exosome-mediated bone metastasis. This case underscores the value of high-throughput miRNA sequencing in identifying functional exosomal miRNAs and their downstream targets in disease progression.
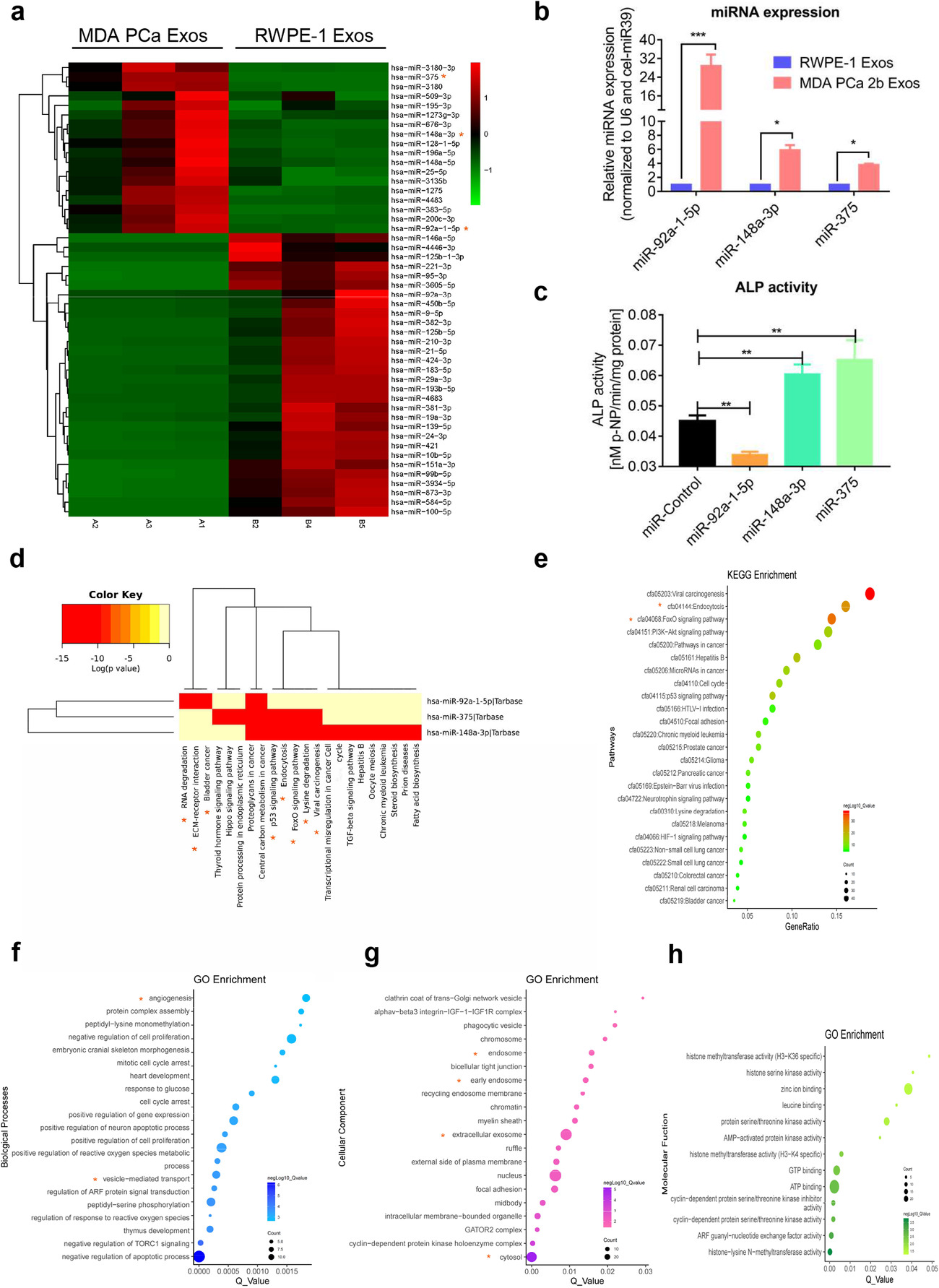
Figure 3. miRNAs Transferred by Prostate Cancer-Derived Exosomes Regulate Bone Homeostasis
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Raw Data Files
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Customized Report Tailored to Client's Requirements
For more information about our microRNA Sequencing Service, please contact us. Our technical specialists are available to provide a free business assessment.
Related Services
Single-Cell miRNA Sequencing Service
How to order?







