mRNA Sequencing Service
- Denaturation – heating the sample to separate the double-stranded cDNA
- Annealing – allowing primers to bind to specific target sequences
- Extension – using DNA polymerase to synthesize new DNA strands from the primers
- Cells: ≥ 1 × 107
- Fresh plant tissue: ≥ 500 mg
- Animal tissue: ≥ 300 mg
- Blood: ≥ 2 mL
- FFPE sections: ≥ 10 unstained slices (no HE staining)
- RNA Samples: Total RNA ≥ 3 μg
- Store at –80 °C and ship on dry ice
mRNA transcriptome sequencing, also known as messenger RNA sequencing (mRNA-Seq), RNA sequencing (RNA-Seq), or transcriptome profiling, is a powerful technique for analyzing messenger RNA molecules within cells. This method enables researchers to determine which genes are actively being expressed in a specific cell or tissue at a given time, and to quantify their expression levels. By applying high-throughput sequencing platforms, mRNA-Seq provides a deep, comprehensive view of gene expression under defined biological conditions. It allows for the identification of gene regulatory networks, detection of disease-associated gene expression patterns. As a result, mRNA-Seq plays a critical role in modern biomedical research, offering valuable data to support precision medicine, functional genomics, and gene therapy development.
MtoZ Biolabs provides professional mRNA Sequencing Services to support transcriptome research across a wide range of biological and biomedical applications. Our mRNA Sequencing Service enables high-throughput, unbiased analysis of gene expression by sequencing the complete set of mRNA transcripts in cells or tissues. Using optimized library preparation protocols and advanced Illumina sequencing platforms, we deliver accurate, reproducible, and customizable transcriptome profiles to help researchers uncover gene regulation patterns, identify biomarkers, and explore disease mechanisms with confidence.
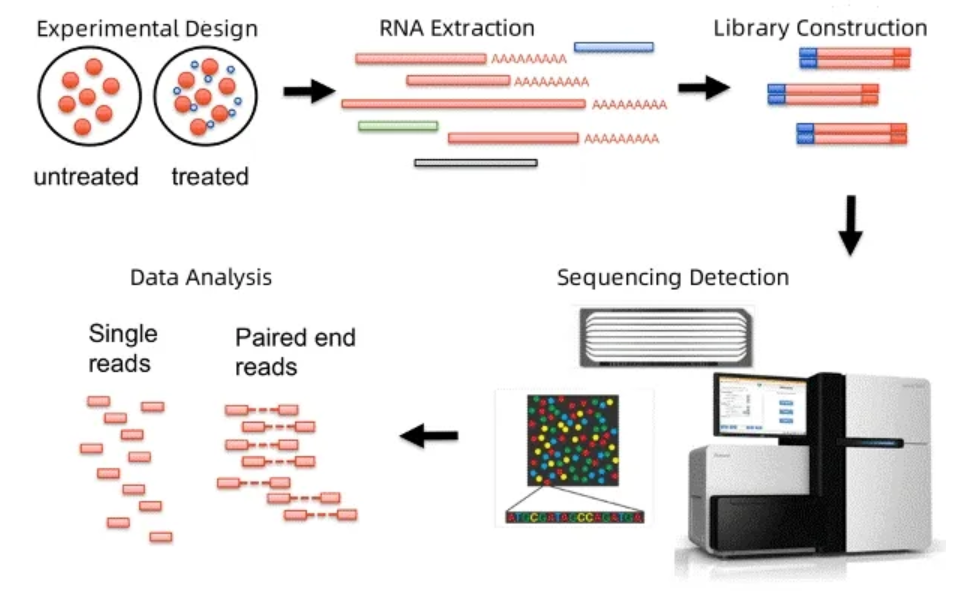
Figure 1. Workflow for mRNA Sequencing Service
Technical Principles
The core principle of mRNA sequencing is based on reverse transcription polymerase chain reaction (RT-PCR) combined with second- or third-generation sequencing technologies. The process begins by extracting mRNA from cells, converting it into complementary DNA (cDNA), and then subjecting the cDNA to high-throughput sequencing.
mRNA sequencing involves two main steps:
1. Reverse Transcription (RT)
In this step, reverse transcriptase synthesizes complementary DNA (cDNA) from the RNA template. This process requires a primer—typically either random hexamer primers or oligo(dT) primers that bind to the poly-A tail of mRNA. Reverse transcriptase then extends from the primer to create a cDNA strand complementary to the original mRNA.
2. Polymerase Chain Reaction (PCR)
Following cDNA synthesis, PCR amplification is performed to generate sufficient DNA for sequencing. PCR consists of three main steps:
These steps are repeated in cycles to exponentially amplify the cDNA. The resulting amplified DNA is then used to construct sequencing libraries for downstream analysis using high-throughput sequencing platforms such as Illumina or third-generation systems.
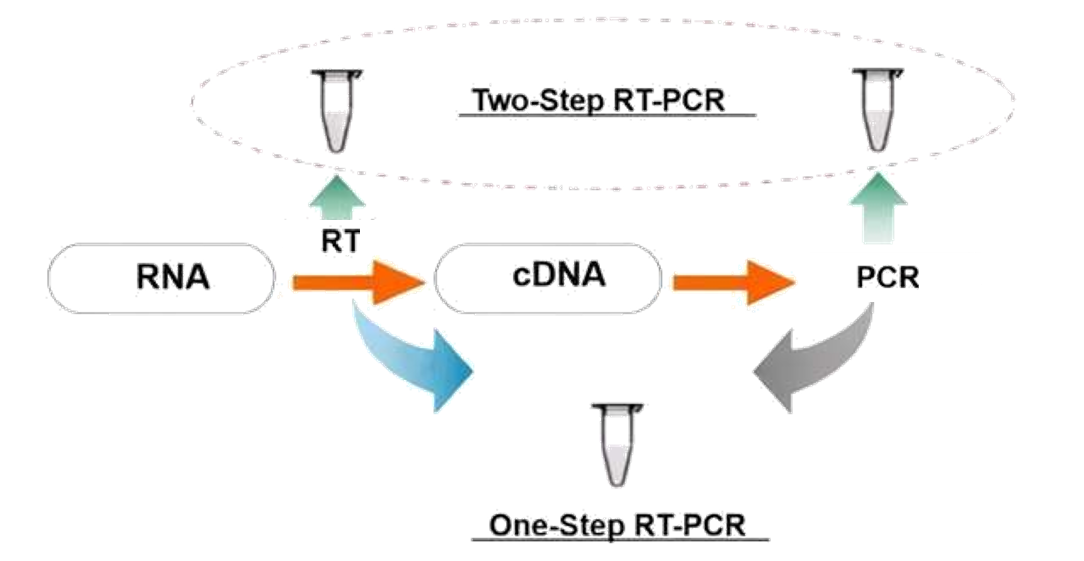
Figure 2. Principle of mRNA Sequencing Service
Service Advantages
☑️High Throughput: Enables simultaneous quantification of thousands to millions of transcripts.
☑️High Sensitivity: Detects low-abundance transcripts, ideal for studying rare gene expression.
☑️Wide Dynamic Range: Accurately measures genes with vastly different expression levels.
☑️Unbiased Detection: No need for probes or antibodies; captures all transcripts without bias.
☑️Multifunctional Output: Supports expression quantification, novel transcript discovery, splicing, and fusion detection.
☑️Broad Sample Compatibility: Applicable to diverse sample types, including cells, tissues, blood, and urine.
☑️Cross-Sample Comparability: Standardized workflows allow consistent analysis across conditions and replicates.
☑️Data Richness: Generates large datasets suitable for differential expression, functional, and pathway analysis.
☑️Streamlined Workflow: Simplified sample preparation and analysis compared to traditional methods.
Applications
1. Gene Expression Regulation Research
Analyze transcriptional dynamics across different cell types, developmental stages, or physiological states to uncover gene regulatory networks and signaling pathways.
2. Disease Mechanism and Biomarker Discovery
Identify aberrantly expressed genes associated with diseases such as cancer, autoimmune disorders, and neurological conditions, supporting early diagnosis, subtyping, and prognosis.
3. Drug Discovery and Development
Explore drug mechanisms of action, validate therapeutic targets, evaluate efficacy, and assess off-target effects or toxicity at the transcriptome level.
4. Plant and Agricultural Genomics
Investigate gene expression patterns related to important agronomic traits such as stress resistance, yield, and quality to support molecular breeding and crop improvement.
5. Environmental and Microbial Studies
Examine transcriptional responses of microorganisms or host organisms under environmental stress, pollutant exposure, or ecosystem shifts.
Sample Submission Suggestions
1. Sample Types
We accept various sample types, including but not limited to:
2. Storage & Shipping
*Note: Please contact us before shipping for any special requirements or pre-analysis consultation.
Case Study
Propionic acidemia (PA) is a rare genetic metabolic disorder caused by mutations in the propionyl-CoA carboxylase (PCC) genes, leading to impaired metabolism and toxic accumulation of organic acids. mRNA-3927 is an investigational mRNA therapeutic encapsulated in lipid nanoparticles (LNPs) containing polyethylene glycol (PEG), which enhances targeted delivery to hepatocytes by limiting phagocytic uptake in the liver. The formulation is designed to restore hepatic function by delivering functional PCC subunits via intravenous infusion.
This Phase 1/2 clinical trial utilized an open-label, dose-escalation design to evaluate the safety and preliminary efficacy of mRNA-3927 in patients aged 1 year and older with PA. Participants were divided into five dosing cohorts and received varying intravenous doses of mRNA-3927. The primary endpoints included the incidence and severity of treatment-related adverse events (AEs) and serious adverse events (SAEs). Secondary endpoints involved changes in plasma 2-methylcitrate (2-MC) and 3-hydroxypropionate (3-HP) levels, evaluation of anti-PEG antibodies, and pharmacokinetic profiling of PCCA and PCCB mRNA levels.
This study represents the first-in-human evaluation of an mRNA-based therapeutic for PA. Preliminary findings demonstrated that mRNA-3927 was generally well tolerated and led to reductions in disease-related biomarkers and metabolic decompensation events (MDEs), suggesting promising therapeutic potential for PA patients.
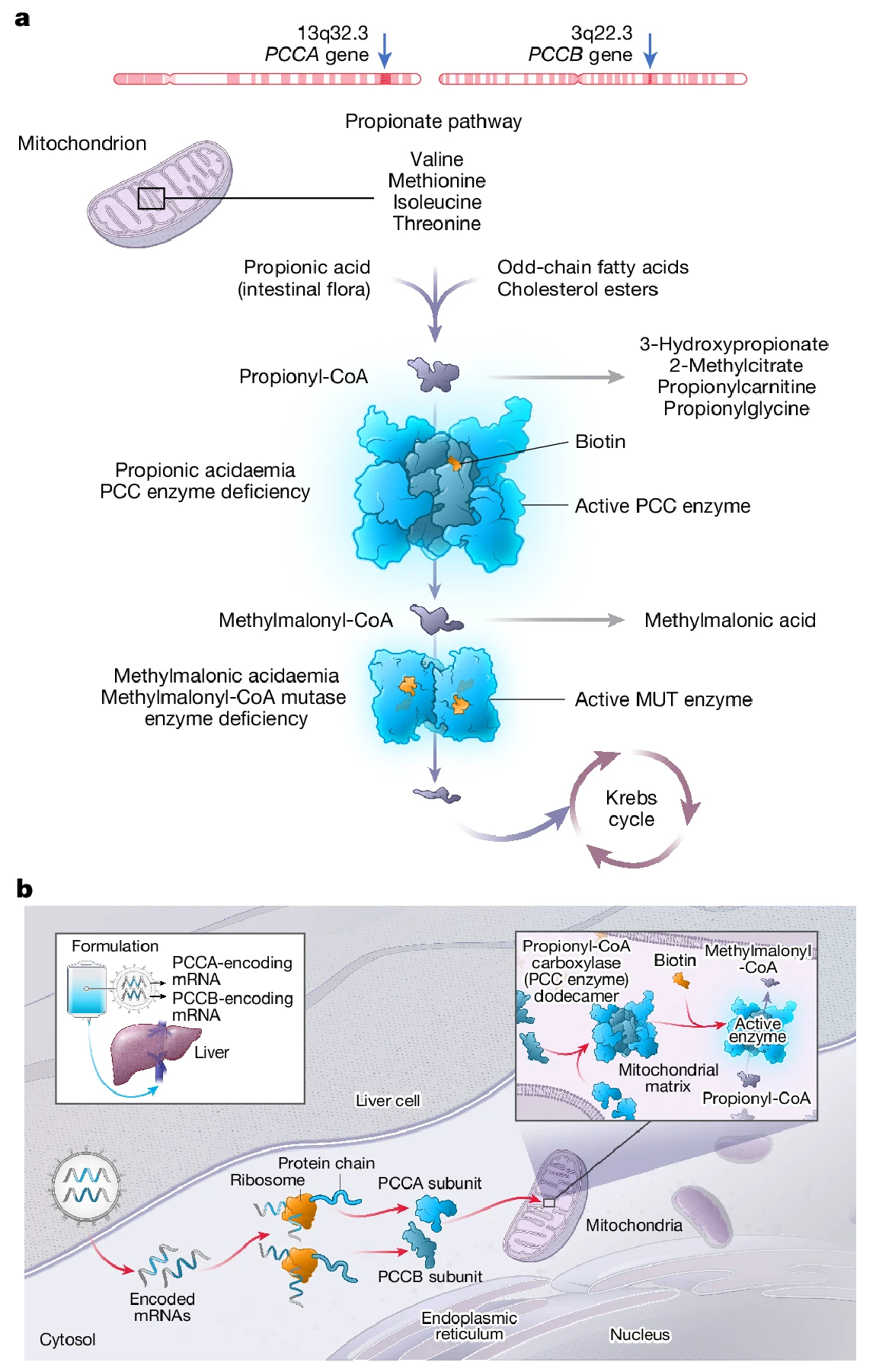
Figure 3. PCCA and PCCB Genes, the Propionate Pathway and the Mechanism of Action of mRNA-3927
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Raw Data Files
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Customized Report Tailored to Client's Requirements
Related Services
Eukaryotic Transcriptome Sequencing Service
How to order?







