Meta-PTMomics Analysis Service
Meta-PTMomics Analysis Service is a systems-level omics analysis service that integrates multiple types of post-translational modifications (such as phosphorylation, acetylation, ubiquitination, methylation, and glycosylation). This service not only identifies various modification sites but also focuses on the cross-talk and cooperative regulation between different modifications, aiming to construct a comprehensive protein modification network map.
Heitel P. Molecules. 2023.
Post-translational modifications (PTMs) are key mechanisms for regulating protein function, stability, localization, and interactions. Common PTMs include phosphorylation, acetylation, ubiquitination, glycosylation, and lipidation. Traditional PTM studies often focus on a single type of modification, making it difficult to fully uncover the synergistic effects and cross-regulation among different PTMs. Meta-PTMomics analysis integrates the identification and quantification of multiple PTMs to provide a panoramic view of protein modification states, facilitating the dissection of complex cellular signaling pathways and disease mechanisms. Meta-PTMomics not only identifies individual modification sites, but also emphasizes the investigation of modification co-occurrence, interactions, and dynamic coordination patterns, serving as a powerful approach for integrated analysis of protein regulatory mechanisms.
MtoZ Biolabs provides Meta-PTMomics Analysis Service using high-resolution mass spectrometry and multidimensional enrichment strategies to comprehensively map protein state changes, regulatory associations, and functional distributions under multiple PTM contexts, offering strong support for exploring complex cellular processes, the molecular basis of diseases, and target intervention strategies.
Analysis Workflow
The Meta-PTMomics Analysis Service workflow at MtoZ Biolabs includes the following steps:
1. Project Design and Sample Preparation
Determine the analysis strategy based on research objectives and provide guidance on sample collection and processing.
2. Multiplex PTM Enrichment
Use targeted enrichment methods to isolate peptide segments with different types of modifications.
3. Mass Spectrometry Data Acquisition
Perform data acquisition using high-resolution mass spectrometry to ensure accurate identification and quantification of modifications.
4. Data Analysis and Interpretation
Integrate data from multiple PTM types to perform functional annotation, pathway analysis, and network construction, delivering in-depth biological insights.
5. Report Delivery
Provide a comprehensive analysis report including modification site information, quantitative results, functional annotations, and visualized charts.
Applications
Meta-PTMomics analysis is widely applied in the following areas:
Signal Pathway Research
Revealing the synergistic roles of multiple post-translational modifications in signal transduction mechanisms.
Disease Mechanism Elucidation
Identifying disease-associated modification patterns and discovering potential biomarkers and therapeutic targets.
Drug Mechanism of Action Studies
Evaluating the impact of drug treatment on protein modification states to support drug development and optimization.
Systems Biology Research
Integrating multi-omics data to construct comprehensive regulatory networks and gain insights into complex biological processes.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
FAQ
Q. Can the Coexistence of Multiple Modifications on the Same Peptide be Identified?
Yes. Our Meta-PTMomics Analysis Service supports the accurate identification and localization of multi-PTM peptides. Leveraging high-resolution mass spectrometry platforms and optimized digestion and enrichment strategies, we can resolve the coexistence patterns of different modifications (such as phosphorylation + acetylation or ubiquitination + methylation) on the same peptide segment, and precisely localize modification sites using site localization scoring.
Q. Is Quantitative Integration of Different Modification Types Supported?
Yes. MtoZ Biolabs can perform cross-modification quantitative integration using unified quantification strategies such as TMT, SILAC, or label-free approaches based on client needs. Through standardized sample preparation and analytical workflows, combined with normalization algorithms and multiple hypothesis testing, we ensure data comparability across different PTM types.
Case Study
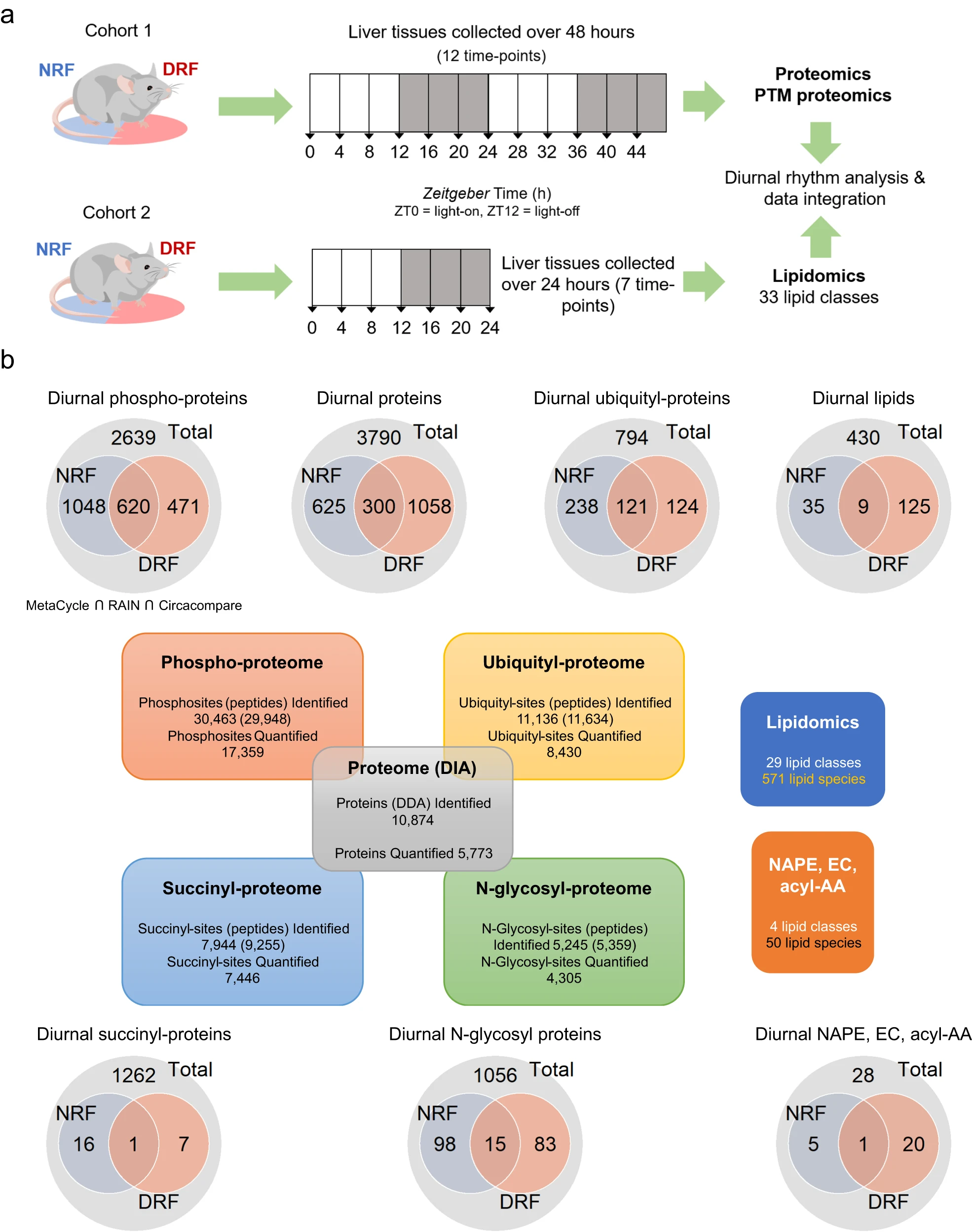
This study performed multi-PTMomics profiling of mouse liver tissue, covering four key types of post-translational modifications: phosphorylation, ubiquitination, succinylation, and N-glycosylation. The results showed that under daytime-restricted feeding (DRF) conditions, phosphorylation exhibited the most pronounced circadian rhythmicity, with approximately 44.8% of phosphorylated proteins displaying rhythmic changes, while succinylation showed the weakest rhythmicity. Notably, the study identified that phosphorylation at Ser971 of the circadian regulator PER2 is modulated by feeding time, suggesting its potential role in hepatic clock regulation. These findings indicate that different feeding schedules can profoundly reshape the rhythmic expression patterns and functional associations of multiple PTMs in the liver.
Huang R. et al. Nat Commun. 2023.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption.
How to order?







