Mass Spectrometry-based m6A Modification Quantitative Analysis Service
Mass Spectrometry-based m6A Modification Quantitative Analysis Service is a high-sensitivity RNA modification quantitative analysis service based on LC-MS/MS. It aims to qualitatively identify and quantitatively assess the presence, abundance, and changes of m6A modifications under different conditions or treatments by digesting RNA into mononucleotides or specific oligonucleotide fragments.
N⁶-methyladenosine (m6A) is one of the most abundant and functional chemical modifications in eukaryotic mRNA. It is also widely found in lncRNA, snRNA, viral RNA, and certain rRNA fragments. m6A modification regulates RNA splicing, export, stability, translation efficiency, and degradation, playing a crucial role in processes such as embryonic development, cell fate determination, tumorigenesis, neurodevelopment, and immune responses.
Traditional m6A analysis methods (e.g., MeRIP-seq) rely on antibody enrichment and high-throughput sequencing. While they enable modification site enrichment across the entire transcriptome, they suffer from issues such as low resolution, poor quantification, and strong dependency on antibody specificity. In recent years, mass spectrometry (MS) technology, with its high resolution, antibody independence, and absolute quantification capability, has become a reliable tool for accurately quantifying m6A modification levels and their dynamic changes.
Leveraging advanced chromatography and mass spectrometry platforms, MtoZ Biolabs offers Mass Spectrometry-based m6A Modification Quantitative Analysis Service providing highly sensitive, antibody-independent quantification of m6A modification levels in RNA samples. This service helps researchers comprehensively assess the dynamic changes of m6A in different biological states, treatment conditions, or disease models, making it suitable for transcriptional regulation mechanism analysis, epitranscriptomics research, and m6A target discovery.
Analysis Workflow
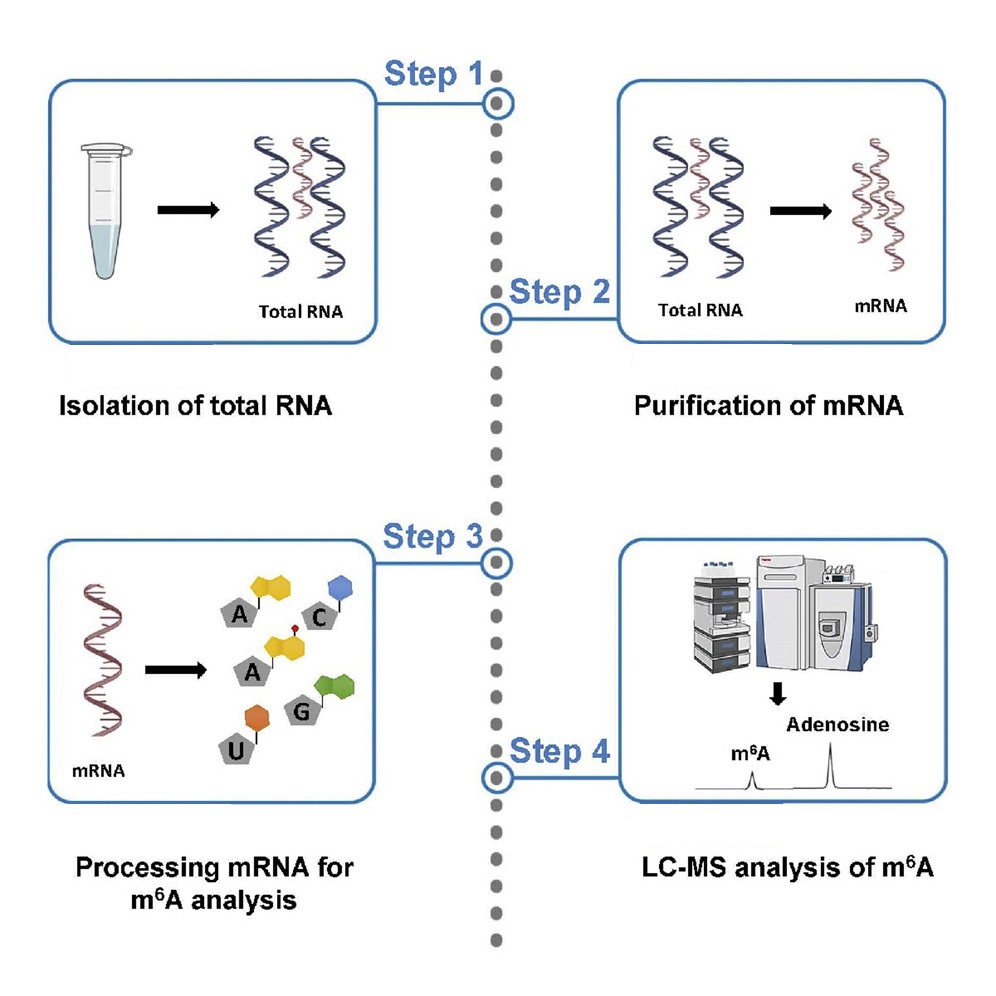
Mathur L. et al. STAR Protoc. 2021.
The main workflow of Mass Spectrometry-based m6A Modification Quantitative Analysis Service is as follows:
1. Sample Preparation
Extract total RNA from the sample and, if required, enrich for mRNA or remove rRNA.
2. Enzymatic Digestion and Nucleotide Preparation
Digest RNA into individual nucleotides and perform purification.
3. LC-MS/MS Detection
Use high-resolution mass spectrometry to detect the nucleotides.
4. Data Processing and Quantitative Analysis
Utilize specialized software for chromatographic peak identification and integration, and calculate m6A concentration or modification ratio using standard curves or internal standards.
5. Result Delivery
Provide comprehensive m6A quantification results, spectra, statistical charts, and a standardized analysis report.
Applications
Application Examples of Mass Spectrometry-based m6A Modification Quantitative Analysis Service:
Cancer Research: Analyze the role of m6A modifications in tumorigenesis and progression, exploring potential biomarkers.
Neuroscience: Investigate the function of m6A in neurodevelopment and neurodegenerative diseases.
Stem Cell and Regenerative Medicine: Explore the regulatory mechanisms of m6A modifications in stem cell differentiation and tissue regeneration.
Plant and Microbiology: Study the role of m6A in plant growth, stress resistance, and microbial metabolism.
Drug Development: Assess the impact of candidate drugs on m6A modification levels, aiding efficacy and toxicity studies.
FAQ
Q. What are the Advantages of Using Mass Spectrometry for Detecting m6A? How does It Differ from MeRIP-Seq?
Mass spectrometry offers advantages such as antibody independence, accurate quantification, and high sensitivity, making it especially suitable for analyzing the total m6A level or the dynamic modifications in specific RNA populations. In contrast, MeRIP-seq is useful for modification mapping across the entire transcriptome, but it has limited resolution, significant antibody bias, and lacks absolute quantification capabilities. Both methods can complement each other.
Q. Does your Service Provide Absolute or Relative Quantification?
MtoZ Biolabs can provide either relative quantification (m6A/A ratio) or absolute quantification (by creating a calibration curve with standards and calculating nucleotide concentrations). If precise molar content is needed, please choose the absolute quantification service and provide additional information to match the internal standard.
Q. What is the Minimum Starting Amount of RNA for Analysis? What are the Quality Requirements?
It is recommended to submit ≥1 µg total RNA per sample, with a concentration of at least 100 ng/µL and good integrity (RIN ≥ 7) to avoid degradation. For rare or clinical samples, please contact us in advance, and we can customize small-volume optimization protocols.
Case Study
This study used LC-MS/MS to quantitatively analyze m6A modification levels in early Drosophila embryos. The results showed that the abundance of m6A modifications in maternal mRNA was significantly higher than in mutant (mettl3 and mettl14 knockout) embryos, indicating that m6A modifications play a key role in maternal mRNA degradation. Further experiments demonstrated that the FMR1 protein preferentially binds to mRNA containing the m6A-modified "AGACU" sequence, and this high-affinity binding depends on the hydrophobic network within the KH2 domain of FMR1. This binding promotes the condensation of FMR1 granules, effectively recruiting unmodified mRNA. As maternal mRNA degrades, FMR1 granules undergo decondensation, allowing for normal embryo development. These findings reveal that m6A modifications regulate the dynamic phase transition of FMR1 granules, facilitating the degradation of maternal mRNA and ensuring proper embryo development.
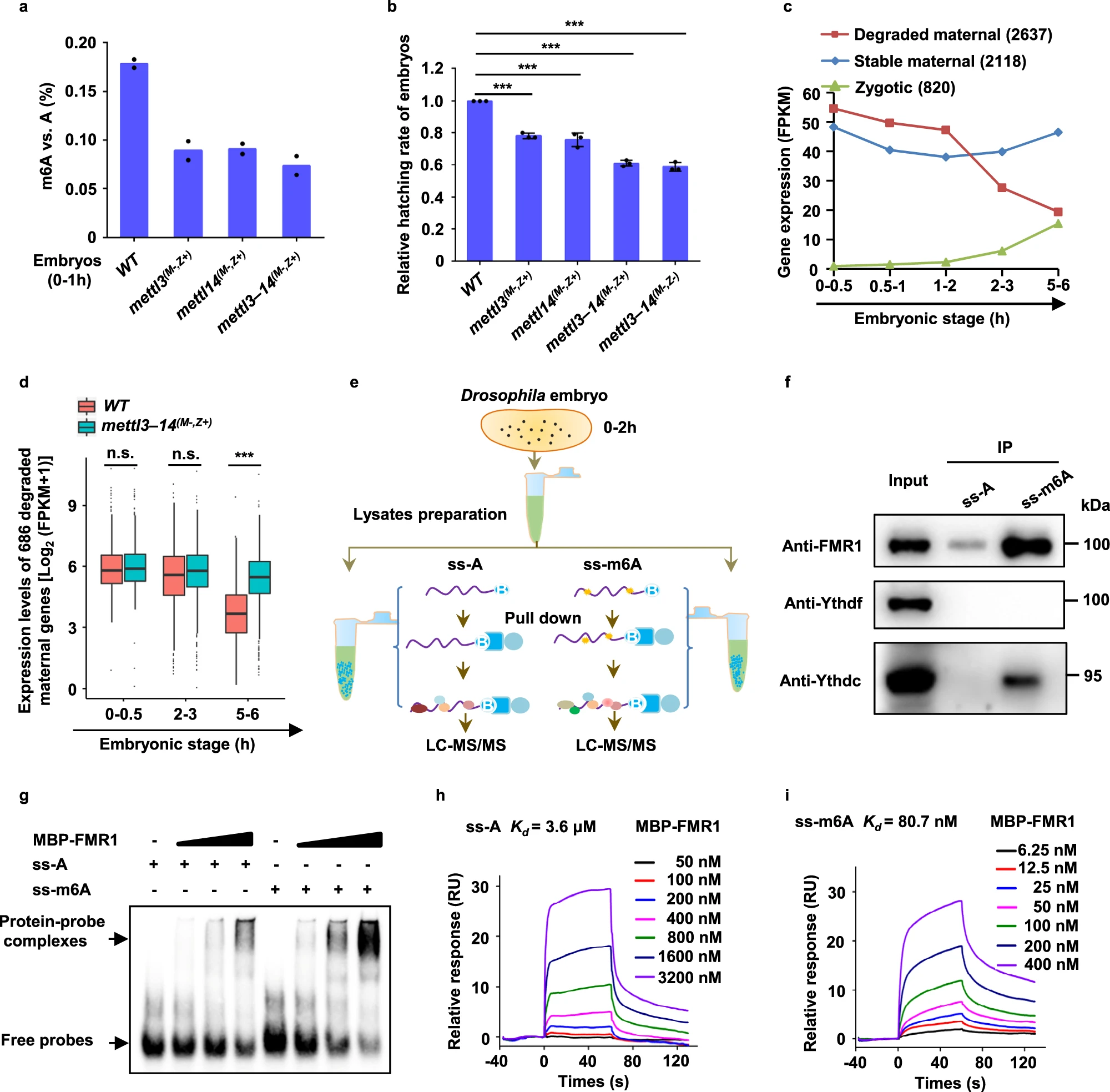
Zhang G. et al. Nat Commun. 2022.
How to order?







