Label-Free vs. Label-Based Quantitative Proteomics: Which One to Choose?
-
A full spectrum of platforms: LFQ, TMT, and DIA
-
Compatibility with diverse sample types (e.g., cells, blood, tissues, exosomes)
-
Extensive experience in differential expression analysis, pathway enrichment, and PPI network construction
-
Professional bioinformatics support for manuscript preparation and project applications
In proteomics research, the choice of quantitative strategy directly influences the resolution, reliability, and breadth of application of the experiment. Given the diversity of sample types and research objectives, researchers often face difficulties in deciding between Label-Free Quantitative Proteomics (Label-Free) and Label-Based Quantitative Proteomics (Labeling, such as TMT or iTRAQ). This article provides a comprehensive comparison of these two major approaches—examining their underlying principles, performance, application scenarios, and cost-effectiveness—to support informed decision-making and help researchers avoid unnecessary trial-and-error.
Fundamental Principles: Key Differences Between Quantification Approaches
1. Label-Free Quantitative Proteomics (LFQ)
LFQ does not involve any chemical or metabolic labeling. Instead, peptide abundance is inferred directly from mass spectrometric data, either by signal intensity or by spectral count. Each sample is analyzed in a separate LC-MS/MS run, and relative expression levels are derived through computational alignment and normalization.
(1) MS1 Intensity-Based Approach: Relies on the chromatographic peak area of peptide ions
(2) Spectral Counting Approach: Based on the number of MS/MS spectra matched to peptides
2. Label-Based Quantitative Proteomics
This strategy involves tagging samples with isotopic or isobaric labels, enabling co-analysis, co-ionization, and co-detection within a single run—thus minimizing technical variation. Commonly used methods include:
(1) TMT/iTRAQ: Isobaric labeling techniques allowing simultaneous comparison of up to 16 samples
(2) SILAC: Incorporates stable isotope labels during cell culture to ensure biological consistency
MtoZ Biolabs offers comprehensive LFQ and TMT-based quantification services tailored to various experimental needs, covering a wide range of sample types including human, animal, plant, and bacterial specimens.
Performance Comparison: Which Is More Accurate? Which Has Higher Throughput?
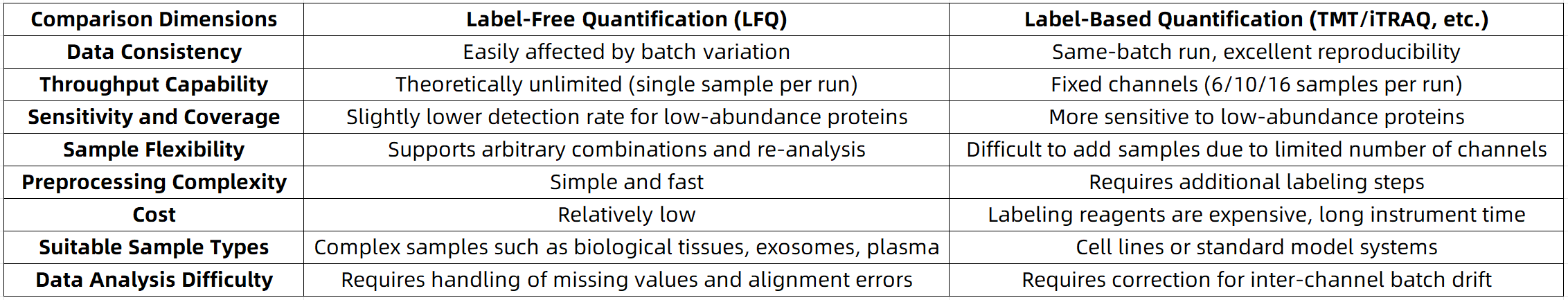
Selection Guide: How to Choose Based on Different Research Goals?
For large-scale screening of protein expression in clinical samples—such as proteomic profiling of tumor tissues, blood, or other body fluids—label-free quantitative proteomics (LFQ) is generally the preferred approach. These studies often involve a wide variety and large volume of samples that require batch-wise collection and sequential analysis. The advantages of LFQ, including high throughput, cost efficiency, and flexible sample handling, make it particularly well-suited for such exploratory research.
For studies aiming to elucidate drug mechanisms or validate molecular targets—for example, analyzing protein expression differences in cell lines under various treatments—label-based quantitative proteomics strategies such as TMT or iTRAQ are recommended. These methods allow for the simultaneous quantification of multiple treatment conditions within a single mass spectrometry run, minimizing batch effects while offering enhanced sensitivity for low-abundance proteins. This makes them especially suitable for pharmacological research demanding high reproducibility and precision.
When the objective is to investigate biological pathways or construct regulatory networks, the choice of strategy can be adapted according to sample throughput. For smaller sample sizes, TMT provides a more consistent and reliable data baseline. In contrast, for large-scale or longitudinal sample sets, LFQ combined with data-independent acquisition (DIA) is recommended to achieve a comprehensive and reproducible protein expression landscape.
Latest Trends: Integration of Quantitative Proteomics Technologies
1. DIA-LFQ: Integration of DIA-Based Data Acquisition and LFQ Quantification
This hybrid approach enhances both data completeness and consistency in differential protein identification, representing an advanced version of LFQ.
2. LFQ + TMT Hybrid Strategy
A subset of samples can be labeled with TMT for in-depth analysis, while the remaining samples are analyzed using LFQ for large-scale expansion. This strategy leverages both high-resolution depth and broad-scale coverage.
3. Multi-Omics Integration (Proteomics + Transcriptomics)
Regardless of whether LFQ or TMT is used, the ultimate goal is to integrate multi-dimensional data to gain deeper insights into biological systems and their regulatory mechanisms.
MtoZ Biolabs: Enabling the Right Proteomics Strategy for Your Research
We offer comprehensive quantitative proteomics solutions tailored for scientific research, including:
Whether you're new to proteomics or initiating a large-scale research project, MtoZ Biolabs provides the technical expertise and strategic guidance to help you select the most appropriate approach. Both label-free and label-based quantitative proteomics offer distinct advantages—the optimal choice depends on your study objectives, sample characteristics, and available resources. A technically proficient service provider can ensure robust data generation, reliable results, and efficient research progress.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







