Label-Free DIA Quantitative Proteomics
Label-Free DIA Quantitative Proteomics is a high-throughput, high-sensitivity, and highly reproducible protein quantification method that integrates label-free quantification (LFQ) strategies with data-independent acquisition (DIA) technology. This approach does not rely on isotope labeling but instead utilizes peptide chromatographic peak intensity or peak area for relative quantification while employing DIA’s full-spectrum acquisition strategy to ensure high coverage and reproducibility. By combining the high throughput of label-free strategies with the extensive coverage of DIA technology, Label-Free DIA Quantitative Proteomics is well-suited for large-scale proteomics studies, excelling in applications such as biomarker discovery, disease mechanism research, drug target identification, and cell signaling pathway analysis.
Leveraging an advanced chromatography and mass spectrometry platform, MtoZ Biolabs provides Label-Free DIA Quantitative Proteomics services ensuring high-quality, high-sensitivity, and highly reproducible protein quantification. Contact us to customize your tailored research solution.
Hamzelou S. et al. Critical Reviews in Biotechnology. 2023.
Analysis Workflow
1. Sample Preparation
A standardized protein extraction and trypsin digestion workflow is employed to ensure uniform peptide generation.
Peptides are purified using high-performance liquid chromatography (HPLC).
2. DIA Data Acquisition
Peptides are separated prior to analysis.
A wide-window DIA strategy is applied to capture all peptide ion signals.
3. Data Processing
Spectronaut, DIA-NN, Skyline and other software are used to analyze DIA data in combination with predicted spectral libraries or experimental spectral libraries.
LFQ is performed by integrating chromatographic peak area or peak intensity to calculate the differences in protein expression levels between different samples.
4. Data Analysis and Interpretation
Differential Expression Analysis (DEA): Identifies proteins with significant expression changes.
Functional Enrichment Analysis (GO/KEGG Pathway Enrichment): Examines protein function and biological pathways.
Protein-Protein Interaction (PPI) Network Analysis: Investigates protein regulatory networks.
Applications
Label-Free DIA Quantitative Proteomics has wide applications across various fields, Including but not limited to:
Biomarker Discovery
Used for screening biomarkers associated with cancer, neurodegenerative diseases, and metabolic disorders.
Disease Mechanism Research
Investigates disease-related signaling pathways, analyzing dynamic protein changes and their regulatory mechanisms in physiological and pathological states.
Drug Development
Label-Free DIA Quantitative Proteomics can be used in drug action mechanism research, drug target screening, drug efficacy evaluation and other fields.
Cell Signaling Pathway Studies
Analyzes protein interaction networks under different physiological conditions, uncovering the regulatory mechanisms of signaling pathways.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Label-Free DIA Quantitative Proteomics platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Label-Free DIA Quantitative Proteomics data, providing clients with a comprehensive data report.
4. High Throughput and Cost-Effective: Eliminates the need for expensive isotope labeling. Compared to TMT/iTRAQ labeling methods, it offers higher sample throughput, making it ideal for large-scale proteomics studies.
5. Enhanced Detection of Low-Abundance Proteins: The full-scan DIA acquisition mode ensures that all peptide ions are collected. Combined with deep-learning-based predicted spectral libraries, it optimizes data processing and improves the detection rate of low-abundance proteins.
6. High Quantitative Accuracy and Reproducibility: Label-Free DIA overcomes the stochastic sampling limitations of DDA-based approaches, resulting in higher data reproducibility.
FAQ
Q. How to Choose the Best Quantification Method (Peak Area vs. Peak Intensity)?
In Label-Free DIA quantification, Peak Area and Peak Intensity are two commonly used quantification strategies, each with distinct advantages.
Peak Area (Chromatographic Peak Area):
Suitable for complex samples and proteins with a wide dynamic range.
Less affected by transient signal fluctuations, offering higher quantification reproducibility.
Recommended for studies with large protein abundance variations, such as plasma proteomics.
Peak Intensity (Chromatographic Peak Intensity):
Ideal for short chromatographic gradients and fast scanning modes, improving data processing efficiency.
More stable for high-abundance protein quantification but more sensitive to low-abundance protein signals.
Suitable for applications requiring rapid detection, such as single-cell proteomics.
For large-scale protein quantification, Peak Area is generally preferred over Peak Intensity as it provides higher quantification stability and reproducibility.
Q: How to Reduce False Matches in DIA Data Analysis?
DIA data analysis is susceptible to fragment ion complexity, which can lead to an increase in false matches. Optimization strategies include:
1. Use High-Quality Spectral Libraries: Combine experimental DDA spectral libraries with deep-learning-based predicted spectral libraries to enhance peptide matching accuracy.
2. Strictly Control FDR (False Discovery Rate): Set a 1% FDR threshold at both the peptide and protein levels, and apply q-value optimization to reduce false positives.
3. Optimize Data Processing Software Parameters: Utilize DIA-NN, Spectronaut, and Skyline, adjusting scoring weights to improve true match rates.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study utilized Label-Free DIA Quantitative Proteomics to analyze the PLK1 (Polo-like kinase 1)-dependent G2/M phase protein degradation program and identified a novel role of AKAP2 in mitotic cytoskeletal coordination. The results demonstrated that PLK1-regulated protein degradation is crucial for the precise progression of mitosis, while AKAP2 plays a key role in microtubule dynamics and spindle stability. This research not only expands the understanding of PLK1-mediated mitotic regulatory mechanisms but also provides experimental evidence for exploring new cell cycle regulatory factors.
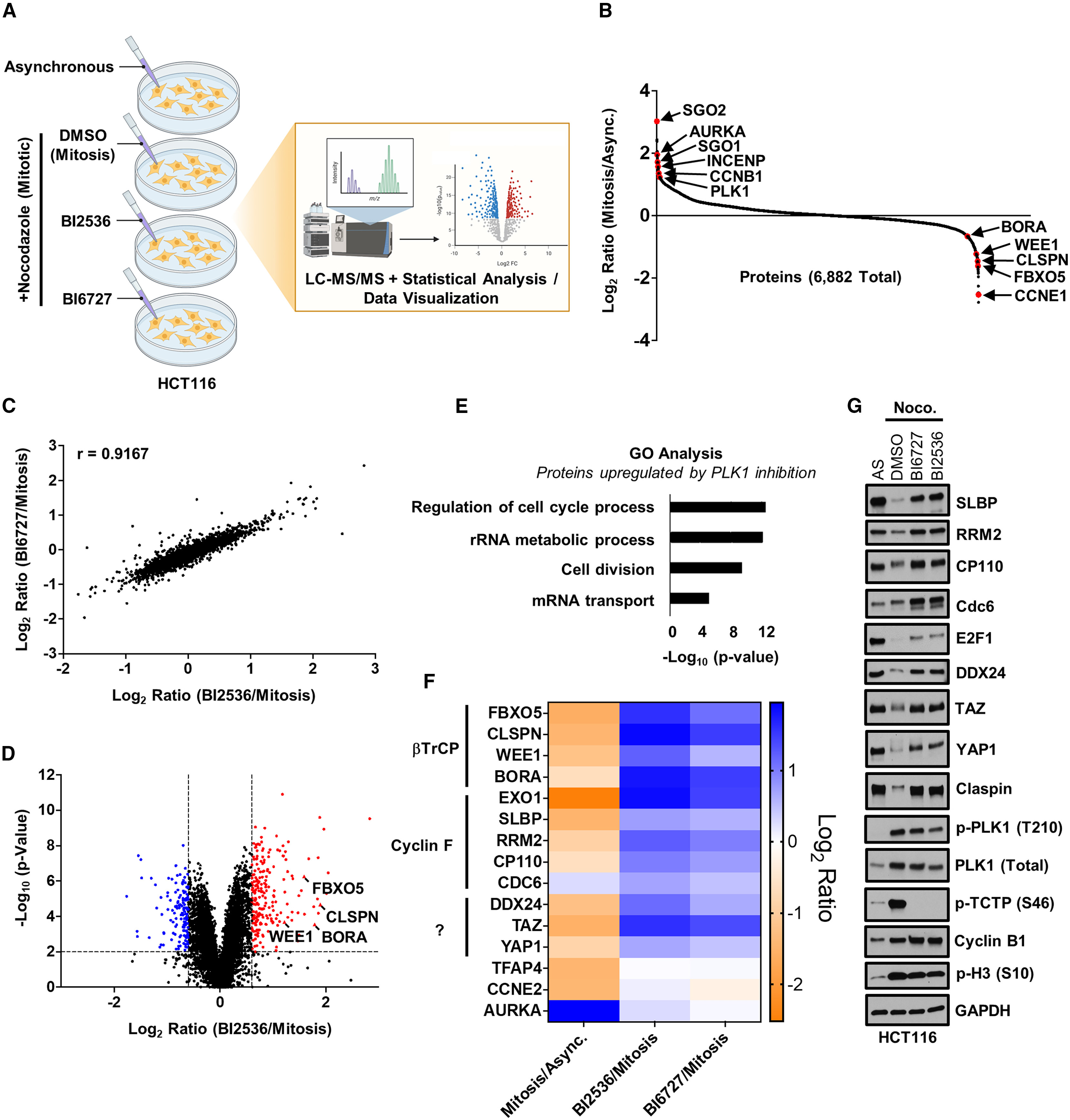
Mouery R D. et al. Cell Reports. 2024.
How to order?







