L-Lactylation Proteomics Analysis Service
L-lactylation proteomics analysis is a high-throughput analytical technique designed specifically to study the post-translational modification involving lactate and proteins. L-lactylation refers to the covalent attachment of lactate to lysine residues in proteins via ester bonds, which can regulate protein function, stability, interactions, and cellular metabolic processes. This modification is closely tied to physiological processes such as cellular metabolism, energy balance, and stress responses, holding significant biological importance.
L-lactylation proteomics analysis service is widely applied in fields including metabolic disorders, cancer research, neurodegenerative diseases, and epigenetics. The technique enables a deeper understanding of how L-lactylation contributes to cellular metabolism and disease progression. It also offers strong technical support for identifying novel drug targets, investigating disease mechanisms, and advancing precision medicine.
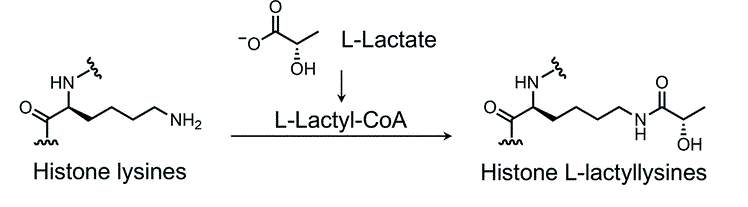
Sun, Y A. et al. Chemical Science, 2022.
Figure 1. Chemical Structure of Histone Lysine Lactylation.
Services at MtoZ Biolabs
Based on high-resolution mass spectrometry (LC-MS/MS) technology, MtoZ Biolabs offers advanced L-lactylation proteomics analysis service that utilizes advanced enrichment strategies and bioinformatics tools to deliver accurate identification and quantification data for protein L-lactylation. This service includes sample preparation, specific modification enrichment, high-sensitivity mass spectrometry detection, and comprehensive data analysis. It ensures high-quality modification site information and dynamic data, providing robust technical support for studying the functions and biological significance of protein L-lactylation.
Analysis Workflow
1. Sample Preparation
Extract total protein from cell or tissue samples and enzymatically digest it into peptides suitable for mass spectrometry analysis.
2. Specific Enrichment
Use proprietary enrichment techniques to capture L-lactylation-modified peptides, enhancing detection sensitivity and specificity.
3. Mass Spectrometry Analysis
Employ high-resolution liquid chromatography-mass spectrometry (LC-MS/MS) platforms for high-throughput identification and quantification of L-lactylation modification sites.
4. Data Analysis
Identify modification sites, perform quantitative evaluations, and conduct functional enrichment analysis using bioinformatics software to uncover the role of L-lactylation in protein function and biological processes.
5. Reporting and Interpretation
Generate comprehensive reports that include modification sites, changes in modification levels, and potential biological function annotations, supporting further research and applications.
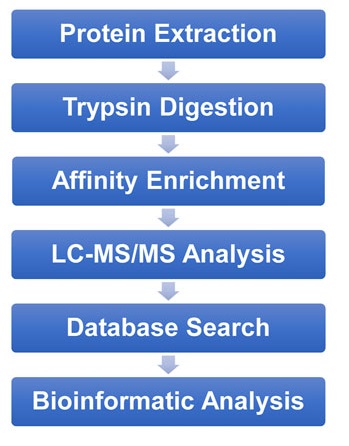
An, D. et al. Frontiers in Genetics, 2022.
Figure 2. The Workflow of L-Lactylation Proteomics Analysis.
Service Advantages
1. High Sensitivity and High Resolution
MtoZ Biolabs has developed an L-lactylation proteomics analysis service platform based on high-resolution mass spectrometry (LC-MS/MS) and optimized enrichment strategies. This ensures accurate identification of low-abundance L-lactylation modifications with high data accuracy and coverage.
2. Versatile Sample Compatibility
The platform supports a wide range of sample types, including cells, tissues, and biofluids, offering flexible options for studying L-lactylation in complex biological systems.
3. Comprehensive Data Support
With advanced bioinformatics tools, we provide modification site identification, quantification of modification levels, and functional enrichment analysis, uncovering the biological functions and underlying mechanisms of L-lactylation.
4. Customized Analysis Solutions
Tailored experimental workflows are designed to align with your research objectives, helping to reveal the role of L-lactylation under various physiological and pathological conditions.
Applications
1. Metabolic Disease Research
L-lactylation is closely linked to metabolic regulation. The L-lactylation proteomics analysis service can be utilized to explore mechanisms of metabolic disorders, such as diabetes and obesity, as well as to identify potential therapeutic targets.
2. Cancer Biology
In cancer research, studying the role of L-lactylation in tumor cell metabolic reprogramming and tumor microenvironment regulation provides insights for discovering novel cancer biomarkers and targeted treatment strategies.
3. Drug Discovery
The specificity and dynamic nature of lactylation make it a promising drug target. By thoroughly analyzing lactylation patterns and their pathological changes, MtoZ Biolabs helps researchers identify novel molecular markers, laying a solid foundation for developing targeted therapies focused on lactylation.
4. Immunology and Inflammation Research
L-lactylation proteomics analysis service can be applied to investigate the impact of L-lactylation on immune cell function and inflammatory responses, uncovering its potential role in immune regulation and autoimmune diseases.
Case Study
1. Lysine L-Lactylation is the Dominant Lactylation Isomer Induced by Glycolysis
This study investigates lysine L-lactylation as the predominant lactylation isomer induced by glycolysis, as well as its biological significance. Using human cell lines and mouse liver tissue, the researchers employed high-resolution mass spectrometry (LC-MS/MS) combined with specific enrichment techniques to systematically identify lysine L-lactylation sites and examine their associations with metabolic environments and gene expression. The results showed that glycolysis-derived L-lactate serves as the primary source of lysine L-lactylation. Compared to D-lactylation, L-lactylation levels were higher and more widely distributed in metabolically active tissues. Functional analyses revealed that L-lactylation is significantly enriched in metabolic enzymes and epigenetic regulators, suggesting it may regulate gene transcription and enzyme activity, thereby modulating cellular function. The study concludes that lysine L-lactylation represents the dominant glycolysis-driven lactylation form, with its formation and dynamic changes tightly linked to metabolic states. This discovery highlights the critical role of L-lactylation in metabolic regulation and gene control, offering new insights for understanding metabolic processes and disease mechanisms.
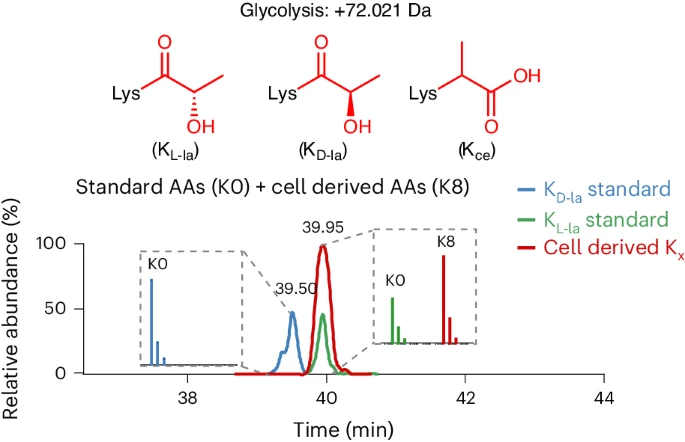
Zhang, D. et al. Nature Chemical Biology, 2024.
Figure 2. The Graphic Abstract of the Study.
Deliverables
1. Experimental Procedures
2. Relevant Mass Spectrometry Parameters
3. Detailed Information on L-Lactylation Proteomics Analysis
4. Mass Spectrometry Images
5. Raw Data
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







