D-Lactylation Proteomics Analysis Service
D-lactylation proteomics analysis is a high-throughput analytical approach dedicated to investigating protein post-translational modifications involving D-lactylation. D-lactylation refers to the covalent attachment of D-lactate to lysine residues on proteins via ester bonds, which can influence protein function, structure, and stability, thereby impacting cellular metabolism and biological processes. This technique typically relies on high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) platforms, employing specific enrichment strategies to precisely identify and quantify modified peptides.
D-lactylation proteomics analysis service is widely applied in fields such as metabolic disorders, cancer, neurodegenerative diseases, and immune-related conditions. By analyzing the dynamic changes of D-lactylation, researchers can uncover its critical roles in energy metabolism, pathway regulation, and disease progression. This information helps identify potential biomarkers and therapeutic targets, offering fresh insights and data support for metabolic regulation and disease mechanism studies.
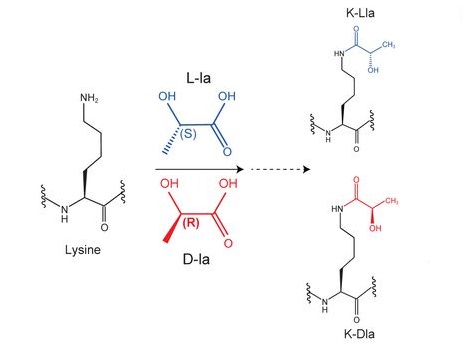
Zang, Y. et al. bioRxiv - Molecular Biology, 2024.
Figure 1. Chemical Structure of Lysine L-Lactylation and Lysine D-Lactylation.
Services at MtoZ Biolabs
Leveraging high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) platforms, alongside specific enrichment techniques and advanced bioinformatics tools, MtoZ Biolabs provides D-lactylation proteomics analysis service that enable the sensitive and high-resolution identification of D-lactylation sites on proteins, as well as accurate quantification of their modification levels. This service encompasses sample preparation, D-lactylation peptide enrichment, mass spectrometry analysis, and bioinformatics data interpretation, ensuring the delivery of high-quality modification site identification, dynamic variation insights, and functional enrichment analysis results. It offers reliable experimental data support for metabolic research, disease mechanism exploration, and new drug development.
Analysis Workflow
1. Sample Preparation
Extract total proteins from cells, tissues, or biological fluids and digest them into peptides, establishing a foundation for subsequent analysis.
2. Specific Enrichment
Use highly specific approaches to selectively capture D-lactylated peptides, enhancing the sensitivity and accuracy of detection.
3. Mass Spectrometry Analysis
Employ high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify and quantify the enriched peptides, thoroughly profiling the modification sites and their distributions.
4. Data Analysis
Utilize bioinformatics tools to quantitatively assess modification sites and perform functional enrichment analysis, uncovering their roles in protein function and biological processes.
5. Results Reporting
Deliver a comprehensive report that includes modification site details, changes in modification levels, and potential functional annotations, providing a solid foundation for further research.
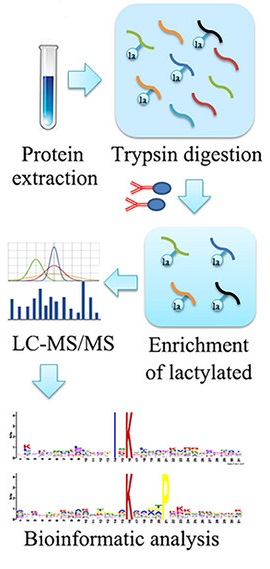
Yang, Y H. et al. Proteomics, 2023.
Figure 2. The Workflow of D-Lactylation Proteomics Analysis.
Service Advantages
1. High Sensitivity and Precision
Using a high-resolution mass spectrometry platform, MtoZ Biolabs has developed a D-lactylation proteomics analysis service platform that ensures accurate identification of D-lactylation sites, making it suitable for detecting low-abundance modifications.
2. Versatile Sample Support
Compatible with various sample types, including cells, tissues, and bodily fluids, the platform accommodates a wide range of research needs.
3. Comprehensive Data Analysis
We deliver modification site identification, quantitative analysis of modification levels, and functional enrichment studies, shedding light on the biological roles of D-lactylation.
4. Customized Experimental Design
Tailored experimental plans are created based on clients’ research objectives and sample types, aiding investigations into the mechanisms of D-lactylation in metabolic regulation and disease development.
Applications
1. Metabolic Disease Research
D-lactoylation modification may have different modification distribution and metabolic pathway in different metabolic States, which is suitable for studying specific metabolic enzyme regulation and key pathways in metabolic diseases.
2. Cancer-Related Signaling Pathway Analysis
D-lactylation proteomics analysis service can identify specific modification patterns in tumor cells, offering insight into unique targets involved in cancer metabolic reprogramming and drug resistance.
3. Epigenetics and Gene Expression Regulation
D-lactylation proteomics analysis service explores the association of D-lactylation with specific transcription factors or histone modifications, uncovering its unique role in regulating gene expression and epigenetic mechanisms.
4. Immune Regulation and Inflammatory Response
D-lactylation may modulate distinct immune cell signaling pathways and the expression of inflammatory mediators, making it valuable for studying the regulatory mechanisms underlying immune-related diseases.
Case Study
1. D-lactate derived from intestinal bacteria drives lysine D-lactylation to modulate transcription in liver cells
This study investigates how the gut bacterial metabolite D-lactate regulates transcriptional activity in liver cells by driving lysine D-lactylation. The research involved human liver cell lines and mouse models, employing methods such as gut microbiota transplantation, metabolite detection, gene expression analysis, and high-resolution mass spectrometry (LC-MS/MS) to identify and quantify modification sites. Results revealed that D-lactate produced by gut bacteria induces lysine D-lactylation in liver cells, significantly impacting the transcriptional regulation of various metabolic and inflammatory signaling pathways. Functional analysis showed that lysine D-lactylation modifies epigenetic regulatory factors, further influencing hepatic gene expression patterns and physiological functions. The study concludes that the interaction between microbial metabolites and host epigenetic regulation offers valuable insight into the mechanisms underlying metabolic diseases and paves the way for innovative therapeutic approaches.
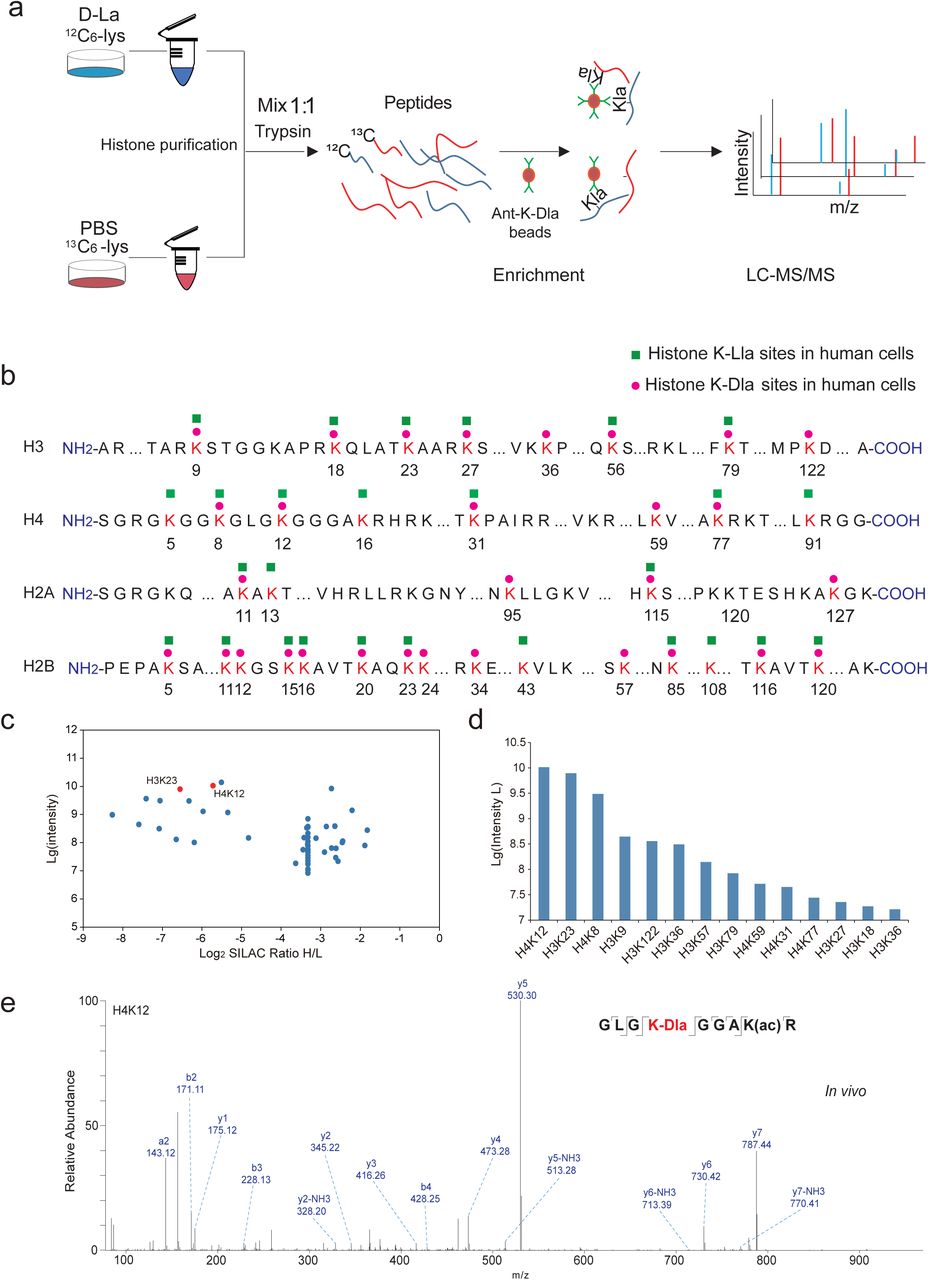
Zang, Y. et al. bioRxiv - Molecular Biology, 2024.
Figure 3. The landscape of histone D-lactylation triggered by D-la.
Deliverables
1. Experimental Procedures
2. Relevant Mass Spectrometry Parameters
3. Detailed Information on D-Lactylation Proteomics Analysis
4. Mass Spectrometry Images
5. Raw Data
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







