Key Differences Between iTRAQ, TMT, and Label-Free Proteomics
-
iTRAQ/TMT: Because labeled samples are combined and analyzed in a single LC-MS/MS batch, inter-batch variability is minimized. This yields higher quantitative consistency and makes these approaches suitable for biomarker discovery and multi-sample comparative studies that require strict quantification accuracy.
-
Label-Free: Although advanced computational algorithms - such as the LFQ implementation in MaxQuant - can enhance quantitative robustness, inter-experiment batch effects remain a major source of variability and can influence quantitative precision.
-
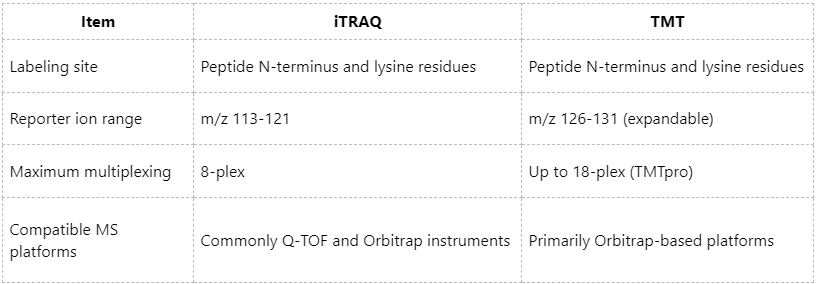
iTRAQ: Typically available in 4-plex or 8-plex formats, suitable for moderate-throughput studies with moderate labeling costs.
-
TMT: Supports up to 18-plex (TMTpro), greatly increasing multiplexing capacity and enabling efficient comparison across large sample cohorts.
-
Label-Free: In principle allows arbitrary sample sizes; however, longer analysis times and increased standardization requirements may offset this advantage.
-
iTRAQ/TMT: Requires additional chemical labeling steps and experienced operators. The LC-MS/MS platform must also support high-resolution and highly reproducible MS/MS acquisition.
-
Label-Free: Sample preparation is relatively straightforward and is often favored for budget-limited projects or laboratories with limited technical experience.
In modern proteomics research, selecting an appropriate quantification strategy is crucial. With advances in mass spectrometry, iTRAQ (Isobaric Tags for Relative and Absolute Quantitation), TMT (Tandem Mass Tag), and label-free quantification have emerged as three mainstream approaches that are widely applied in studies of disease mechanisms, drug target screening, and clinical biomarker discovery. Although all three approaches rely on high-resolution mass spectrometry platforms, they differ substantially in labeling strategy, quantitative accuracy, sample throughput, and data processing workflows.
Comparison of Basic Principles of the Three Quantification Strategies
1. iTRAQ and TMT: Isobaric Labeling Technologies
iTRAQ and TMT are both isobaric labeling strategies. In these approaches, peptides derived from different samples are labeled with isobaric chemical tags of identical precursor masses; as a result, they are indistinguishable at the MS1 level. Upon MS2 fragmentation, distinct reporter ions are released and used for relative quantification across samples.
2. Label-Free Quantification: Intensity-Based Quantification Without Chemical Labels
Label-free quantification (LFQ) does not require chemical tagging. Instead, relative protein abundance is inferred through direct comparison of peptide ion intensities (peak intensity) or spectral counts across different samples.
This approach offers simplicity, no label-related costs, and no inherent limitations on sample number; however, it places strict demands on experimental reproducibility, LC-MS stability, and downstream data normalization procedures.
Core Differences: From Quantitative Accuracy to Application Flexibility
1. Quantitative Accuracy and Data Consistency
2. Sample Throughput and Cost Considerations
3. Experimental Workflow and Technical Barrier
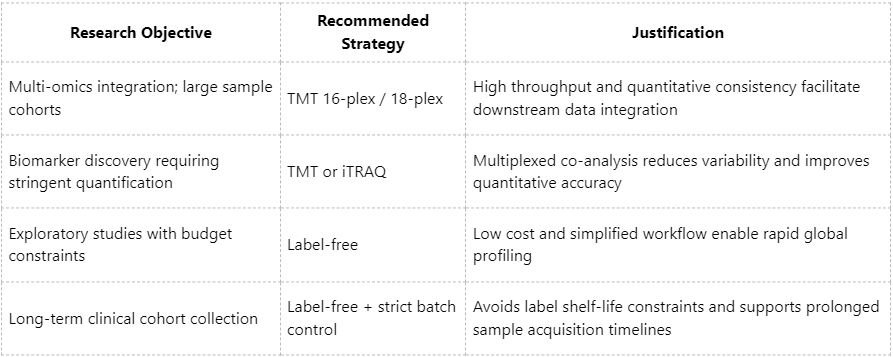
Recommended Application Scenarios: Selecting an Appropriate Quantification Strategy
Technological Advances and Future Trends
Recent improvements in TMT technology - including the TMTpro 18-plex format - have enabled large-scale cohort studies involving multiple disease subtypes or longitudinal time points. In parallel, algorithmic advances such as real-time database search (RTS) and intensity-based enhancement frameworks (e.g., DIA coupled with iRT calibration) have substantially improved the quantitative stability of label-free approaches.
Looking forward, hybrid quantification strategies (e.g., TMT discovery combined with DIA-based validation) are expected to become increasingly prevalent in systems biology research, compensating for limitations of individual methods and enabling more comprehensive and accurate proteome profiling.
Overall, iTRAQ, TMT, and label-free quantification each offer distinct advantages and suitable application contexts. There is no universally optimal solution; instead, the most appropriate strategy depends on research goals, sample characteristics, budget constraints, and instrument capabilities. MtoZ Biolabs provides a full spectrum of proteomics quantification services - including iTRAQ, TMT, and label-free workflows - supported by extensive mass spectrometry expertise and dedicated bioinformatics analysis capabilities to assist researchers in advancing more confidently and effectively in proteomics research.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







