iTRAQ/TMT-Based PTM Quantitative Analysis Service
- Report Group: Emits unique signature ions during MS/MS fragmentation.
- Balance Group: Ensures identical total mass across all tags, enabling co-elution of labeled peptides.
- Reactive Group: Covalently binds to peptide N-termini and lysine side chains.
Post-translational modifications (PTMs) are pivotal regulators of protein function, influencing cellular signaling, enzymatic activity, and disease mechanisms. Over 400 PTMs have been identified, including phosphorylation, ubiquitination, and acetylation, each modulating protein behavior in dynamic biological contexts. Despite their importance, PTMs are often low-abundance and transient, posing challenges for detection and quantification. Traditional label-free approaches, while useful, often lack the sensitivity and consistency needed for low-abundance PTM peptides across multiple samples. To address these limitations, MtoZ Biolabs provides iTRAQ/TMT-Based PTM Quantitative Analysis Service which utilizes cutting-edge mass spectrometry (MS) and isobaric tagging technologies to deliver high-throughput, accurate PTM profiling. This service empowers researchers to uncover novel biomarkers, decode disease pathways, and accelerate drug discovery.
Technical Principles
The service utilizes iTRAQ (Isobaric Tags for Relative and Absolute Quantitation) and TMT (Tandem Mass Tags), two gold-standard isobaric labeling techniques designed for multiplexed quantitative proteomics. These technologies enable simultaneous quantification of PTM-bearing peptides across multiple biological conditions in a single mass spectrometry run, increasing throughput, improving quantitation accuracy, and allowing statistically robust comparisons.They rely on the covalent labeling of peptides with isobaric tags that are identical in mass but generate unique reporter ions upon MS/MS fragmentation. Each tag comprises three components:
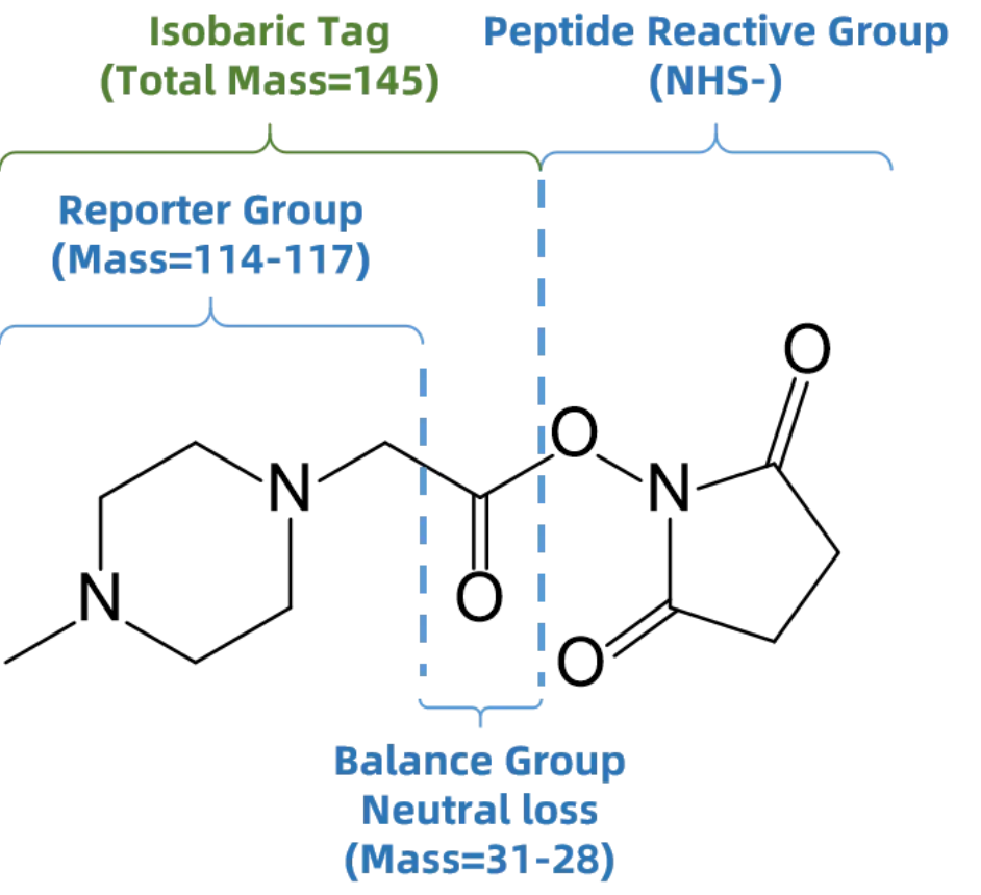
Figure 1. Structure of iTRAQ Tag
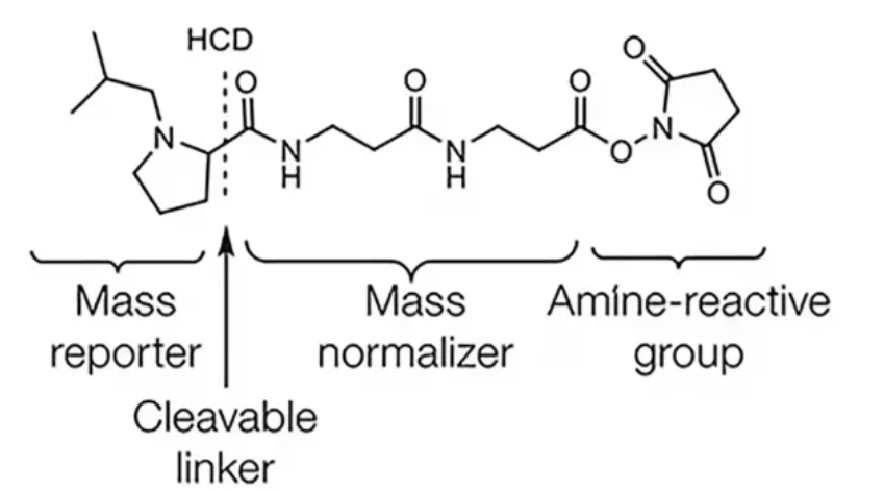
Ege, N. et al. Cell Chemical Biology, 2021.
Figure 2. Chemical Structure Diagram of TMT Label
iTRAQ supports the simultaneous analysis of up to 8 samples, while TMT allows for multiplexing of up to 16 samples. Both techniques can be combined with PTM enrichment strategies—such as phosphopeptide capture or immunoprecipitation—to enable quantitative profiling of low-abundance modifications across complex biological conditions.
Service at MtoZ Biolabs
MtoZ Biolabs' iTRAQ/TMT-Based PTM Quantitative Analysis Service platform integrates:
💠Isobaric labeling with iTRAQ 4-plex/8-plex and TMT 6-plex/10-plex/16-plex
💠PTM-specific enrichment strategies (e.g., TiO₂, IMAC, anti-PTM antibodies)
💠High-resolution LC-MS/MS systems (Orbitrap Fusion Lumos, Q Exactive HF-X)
💠Advanced bioinformatics for site identification, quantification, and functional annotation
Whether you're investigating signaling networks, comparing drug responses, or profiling disease-associated PTM patterns, our service provides a high-throughput, reproducible, and deeply informative solution tailored to your study goals.
Analysis Workflow
1. Sample Preparation: Protein extraction, quantification, and digestion (typically trypsin or Lys-C) from each experimental group.
2. Isobaric Labeling: Each digested sample is labeled with a unique iTRAQ or TMT tag following standard protocols.
3. PTM Enrichment: Enrichment is performed based on the target PTM using antibody-based, chemical, or affinity methods.
4. LC-MS/MS Analysis: Enriched peptides are subjected to high-resolution LC-MS/MS on Orbitrap or Q-TOF systems.
5. Data Analysis: Raw data is processed for identification, quantification, and PTM site localization using advanced analysis software.
6. Bioinformatics Annotation: Quantified PTMs are mapped to pathways, GO terms, interaction networks, and cross-PTM regulation modules.
Griss, J. et al. J Proteome Res. 2019.
Figure 3. Scheme of the Entire Workflow for iTRAQ and TMT
Why Choose MtoZ Biolabs?
☑️Advanced Analytical Platforms
Equipped with state-of-the-art mass spectrometry systems, we ensure high-resolution, high-sensitivity PTM detection and quantification.
☑️Extensive Experience in PTM Proteomics
Our team has deep expertise in iTRAQ/TMT-based proteomics and a wide range of post-translational modifications, including phosphorylation, acetylation, ubiquitination, and methylation.
☑️One-Stop Service
From experimental planning and sample processing to data acquisition, bioinformatics, and reporting, we deliver a fully integrated workflow under one roof—saving you time and ensuring data consistency.
☑️Customizable Experimental Design
We tailor each project to your specific needs, including labeling strategy, PTM enrichment method, instrument parameters, and data analysis depth—ensuring your research goals are met with precision.
☑️Reliable and Reproducible Results
Our optimized protocols and rigorous quality control ensure high reproducibility across technical replicates and biological conditions.
Applications
1. Signal Transduction Pathway Profiling
Quantify phosphorylation, acetylation, or methylation changes in response to signaling stimuli, drug treatments, or genetic manipulation.
2. Disease Mechanism Research
Compare PTM patterns between disease and control samples, responder and non-responder groups, or wild-type and mutant models to uncover modification-driven regulatory mechanisms involved in disease onset, progression, or treatment response.
3. Drug Mechanism and Target Validation
Assess how small molecules, biologics, or inhibitors affect specific PTM sites and pathways.
4. Cross-Talk Between PTMs
Identify competitive or cooperative relationships between PTMs on the same protein or within functional protein complexes.
5. Biomarker Discovery
Identify disease- or treatment-associated PTM changes for translational research or clinical development.
FAQ
Q1: Can I submit partially processed samples?
We can accept digested or labeled peptides, but we prefer to handle the full workflow to ensure consistency and quality control.
Q2: What types of post-translational modifications can be analyzed with this service?
We support a wide range of PTMs including phosphorylation, acetylation, methylation, ubiquitination, SUMOylation, glycosylation, and more. Customized protocols are available for rare or emerging modifications.
Q3: How do I choose between iTRAQ and TMT for my experiment?
Both are isobaric labeling techniques for multiplexed quantification. TMT offers broader multiplexing options (up to 16-plex) and higher MS/MS signal intensity, making it more suitable for large-scale or low-abundance PTM studies. Our team can help you select the best option based on your sample number, target PTM, and instrument compatibility.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Contact us today to discuss your project or request a customized. Let us help you turn complex PTM data into meaningful biological discovery.
How to order?







