Irreplaceability of Full-Length Sequencing:Why Traditional Mass Spectrometry Cannot Achieve Complete Protein Coverage
-
Enzymatic digestion of proteins (e.g., using trypsin) to generate peptide fragments;
-
Mass spectrometric analysis of peptide mass-to-charge ratios and their fragmentation spectra (MS/MS);
-
Computational matching of acquired peptide spectra to reference sequences using database search algorithms (e.g., SEQUEST, Mascot);
-
Inferring the presence of proteins based on the number and sequence coverage of matched peptides.
-
Identification of single nucleotide polymorphisms (SNPs) and amino acid substitutions;
-
Discrimination of structural isomers (e.g., variant light chains in antibodies);
-
Annotation of PTM types and sites (such as phosphorylation, acetylation);
-
Detection of N-terminal/C-terminal processing events, including signal peptide cleavage and N-terminal acetylation.
-
Verifying recombinant protein expression products;
-
Antibody sequencing and humanization analysis;
-
Consistency assessment of therapeutic proteins;
-
Investigating structural and functional features of novel proteins.
In modern proteomics research, mass spectrometry has undoubtedly become a critical tool for advancing the discovery and functional characterization of proteins. From large-scale protein identification to the analysis of post-translational modifications (PTMs), traditional database-dependent mass spectrometry strategies—such as data-dependent acquisition (DDA) and data-independent acquisition (DIA)—offer broad expression profiling capabilities. However, when the focus shifts to deciphering the complete primary structure of a specific protein, researchers frequently encounter a persistent challenge:
“Despite successful measurement and apparent database matches, it remains impossible to reconstruct the full-length amino acid sequence.”
What gives rise to this analytical “blind spot”? And why has protein full-length sequencing become increasingly indispensable? This article systematically explores the limitations of traditional mass spectrometry and the essential value of protein full-length sequencing from three perspectives: mechanistic differences, technical constraints, and application-specific demands.
Basic Logic of Traditional Mass Spectrometry: Database-Based "Puzzle Assembly"
The conventional workflow of proteomics analysis generally follows these steps:
While this approach is high-throughput, broadly applicable, and well-suited for exploratory research, it possesses inherent limitations in reconstructing the complete sequence architecture of a protein.
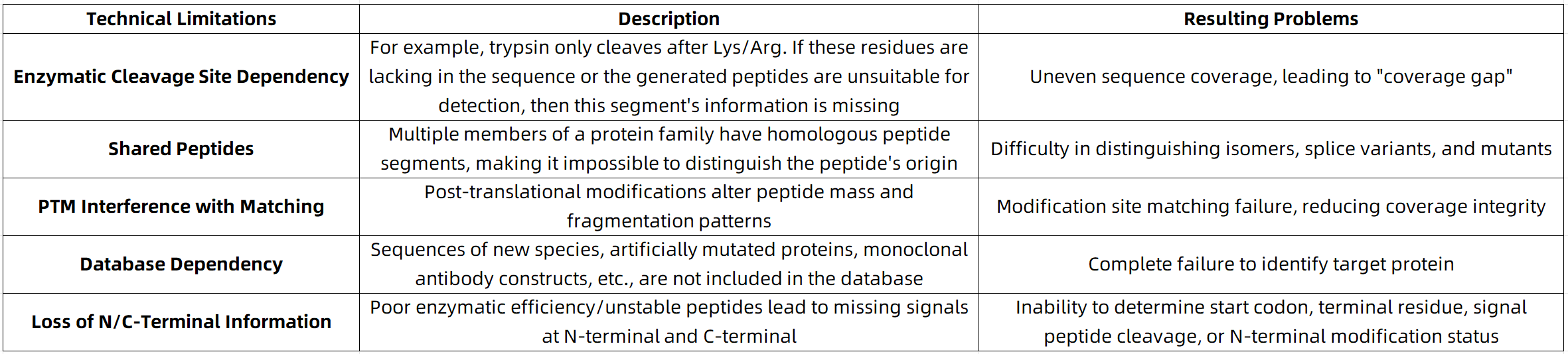
Key Limitations Preventing Full-Length Coverage by Traditional Mass Spectrometry
Fundamental Advantages and Unique Value of Protein Full-Length Sequencing
To address these limitations, de novo protein full-length sequencing has been developed. This approach integrates multi-enzyme digestion, high-resolution mass spectrometry, and AI-driven sequence assembly, offering several distinctive advantages:
1. Database Independence Enables Unbiased Spectrum Interpretation
AI algorithms reconstruct amino acid sequences directly from fragment spectra without referencing existing databases, thereby enabling accurate identification of unknown or mutated proteins.
2. Designed for Complete Sequence Reconstruction
A strategic combination of proteolytic enzymes (e.g., Trypsin, Glu-C, Asp-N, Chymotrypsin) enhances sequence coverage. Redundant peptide fragments provide cross-validation. High-resolution spectra combined with AI-guided assembly support reconstruction of the entire sequence from the N-terminus to the C-terminus.
3. Precise Detection of Mutations, Structural Isomers, and Post-Translational Modifications (PTMs)
4. Enables Comprehensive Validation of the Protein’s Primary Structure
Especially Suitable for:
As proteomics advances toward structure- and function-level resolution, conventional database-dependent strategies fall short of the precision required for molecular-level research. Protein full-length sequencing not only complements traditional techniques but also serves as an indispensable tool for resolving unknown sequences, validating critical regions, and ensuring sequence integrity.
Protein full-length sequencing = Complete coverage + Database independence + Detection of modifications and mutations + N-/C-terminal verification
MtoZ Biolabs combines AI-based spectral interpretation, a comprehensive multi-enzyme digestion system, and high-resolution mass spectrometry to deliver verifiable, traceable, and publication-ready primary protein sequence data, facilitating breakthroughs in structural proteomics.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







