How to Integrate Spatial Proteomics with Multi-Omics Approaches?
The rapid advances in single-cell omics, spatial transcriptomics, and high-throughput mass spectrometry have propelled life-science research into an era of high-dimensional data integration. Conventional proteomics captures changes in protein abundance, yet it often cannot address a fundamental question: where, precisely, are these proteins located within tissues or cells? Spatial proteomics has been developed to fill this gap by enabling visualization and quantitative profiling of protein distributions at subcellular and, in some applications, even single-cell-resolution. Nonetheless, proteins represent only one layer of biological regulation. To elucidate disease mechanisms and identify actionable precision-therapy targets, spatial proteomics is increasingly integrated with other omics modalities, including transcriptomics, metabolomics, phosphoproteomics, and epigenomics, thereby enabling coordinated interpretation across multiple biological dimensions.
Core Technologies and Challenges of Spatial Proteomics
Key technological modalities in spatial proteomics can be broadly grouped as follows:
1. Imaging Mass Spectrometry (IMS)
Techniques such as MALDI-IMS can generate spatial maps of proteins directly from tissue sections. Major strengths include label-free detection and the capacity to profile hundreds of molecular species simultaneously; however, further improvements in spatial resolution and quantitative performance remain important objectives.
2. MS-Based Spatial Proteomics (E.G., LC-MS Coupled with Laser Capture Microdissection)
In this workflow, regions of interest are excised from tissue (e.g., via laser microdissection) and subsequently analyzed by mass spectrometry. When combined with high-resolution imaging, this strategy supports region-resolved proteome profiling and has been widely applied to studies of tumor heterogeneity.
3. High-Plex Imaging Antibody Platforms (E.G., Co-Detection by Indexing, CODEX)
These platforms enable highly multiplexed protein labeling and the construction of single-cell-resolved spatial protein atlases. Their performance, however, is strongly dependent on antibody specificity and the availability of sufficiently large antibody panels.
Key Value of Multi-Omics Integration
Integrating spatial proteomics with other omics layers can expand both research depth and scope in several aspects:
1. Spatially Informed Causal-Chain Inference
Spatial proteomics + spatial transcriptomics: contrasts spatial patterns at the transcriptional and translational levels, facilitating interrogation of post-transcriptional regulatory processes.
Spatial proteomics + spatial metabolomics: delineates the tissue-level spatial distribution of metabolic pathway activities and enables tracking of metabolic reprogramming.
2. Enhanced Biomarker Discovery Through Multi-Dimensional Evidence
Multi-omics integration can reduce background noise and improve biomarker robustness and reproducibility, which is particularly valuable for individualized tumor subtype stratification and investigation of drug-resistance mechanisms.
3. Network Construction from a Systems-Biology Perspective
Integrative algorithms can be used to build protein interaction networks within spatial contexts and to overlay them with transcriptional regulatory or metabolic networks, thereby highlighting core regulatory modules and candidate intervention nodes.
How Can Spatial Proteomics Be Co-Designed with Other Omics Experiments?
Drawing on practical experience, an integrated experimental design may follow a three-step strategy:
Step 1: Spatial Localization Upfront + High-Throughput Screening
Apply spatial proteomics to identify regions of interest (e.g., tumor margin vs. tumor core).
Use bulk RNA-seq or TMT-based quantitative proteomics to nominate candidate biomarkers.
Step 2: Coordinated Interpretation Across Multi-Omics Layers
Integrate spatial transcriptomics (e.g., 10x Visium) with spatial protein maps to construct a spatially resolved functional atlas of tissue domains.
Incorporate metabolomics to identify regions with functional activation or suppression.
Step 3: Validation and Mechanistic Investigation
Leverage phosphoproteomics and epigenomics to further interrogate regulatory mechanisms.
Conduct spatial multi-omics network modeling to pinpoint potential drivers.
Commonly Used Approaches for Integrative Data Analysis
A central difficulty in multi-omics integration is that datasets generated by different platforms can differ substantially in dimensionality, scale, and measurement standards. Widely used approaches include:
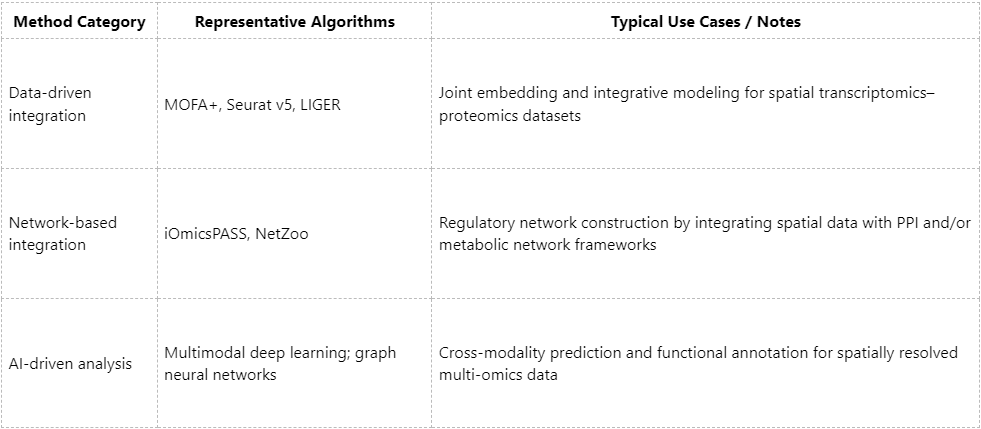
Spatial proteomics is poised to become a key pillar of precision medicine, with utility extending well beyond simply localizing proteins. When integrated with multi-layer omics information, such as transcriptional, metabolic, and epigenetic profiles, it becomes possible to more comprehensively characterize intercellular communication, disease microenvironments, and dynamic pathological processes. At MtoZ Biolabs, we provide end-to-end research support spanning sample pretreatment, spatially resolved processing workflows, mass spectrometry-based measurements, and integrative multi-omics analysis. By delivering rigorous and in-depth interpretations of complex datasets, we aim to help advance your research by enabling critical insights in the spatial dimension.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







