How to Enrich Membrane Proteins: What Are the Most Effective Methods?
- High hydrophobicity: prone to aggregation and precipitation, making them difficult to solubilize in standard buffers;
- Low abundance: typically present in low quantities and often masked by highly abundant soluble proteins;
- Co-extraction with lipids: often co-purified with lipids and cholesterol, thereby interfering with mass spectrometric detection;
- Complex transmembrane topology: multiple membrane-spanning domains hinder complete enzymatic digestion and reduce identification efficiency in MS analysis.
- Lyse cells in an appropriate buffer
- Perform low-speed centrifugation to remove nuclear debris
- Conduct sequential ultracentrifugation to separate plasma, mitochondrial, and other membranes
- Further purification using sucrose density gradients, if required
- Lyse cells with Triton X-114
- Incubate at 20–37°C to induce phase separation
- The aqueous phase retains hydrophilic proteins, whereas the detergent-rich phase concentrates hydrophobic membrane proteins
- Wash and precipitate the proteins for subsequent analysis
- Treat cell lysates with 0.1 M Na2CO3 (pH 11.3) to remove non-membrane-bound proteins
- Collect membrane pellets and solubilize with 1% SDS to release hydrophobic proteins
- Remove residual SDS using approaches such as FASP or S-Trap before mass spectrometric analysis
- Utilizing S-Trap digestion for SDS-containing samples to improve proteolytic efficiency and remove contaminants
- Employing DIA (Data-Independent Acquisition) to minimize missing identifications
- Building spectral libraries for membrane proteins to increase database search accuracy
- Enhancing annotation of hydrophobic peptide sequences to facilitate identification of transmembrane domains
- Focusing on GO terms and pathways related to membrane localization and transmembrane structure during functional analysis
- Highly efficient extraction workflows combining Triton X-114, Na2CO3, and SDS-based methods
- Implementation of Orbitrap Exploris instruments with DIA mode for sensitive identification and quantification
- Integrated analysis including transmembrane domain annotation, pathway enrichment, and data visualization
- Extension to the enrichment and functional characterization of GPCRs, ion channels, and other membrane-associated targets
Membrane proteins play a pivotal role as mediators linking intracellular and extracellular signaling pathways, representing over 30% of the human proteome and encompassing numerous pharmaceutical targets, including GPCRs, ion channels, and receptors. However, due to their high hydrophobicity, poor solubility, and generally low expression levels, membrane proteins are often underrepresented in proteomic analyses. Thus, the membrane protein enrichment is a crucial prerequisite for comprehensive and high-quality studies in membrane proteomics.
Rationale for Membrane Protein Enrichment
Membrane proteins are distributed across various biological membranes, including the plasma membrane, mitochondria, Golgi apparatus, and endoplasmic reticulum, and present several experimental challenges:
Hence, the application of specialized enrichment strategies is fundamental to achieving high-sensitivity proteomic characterization of membrane proteins.
Overview and Comparison of Common Membrane Protein Enrichment Methods
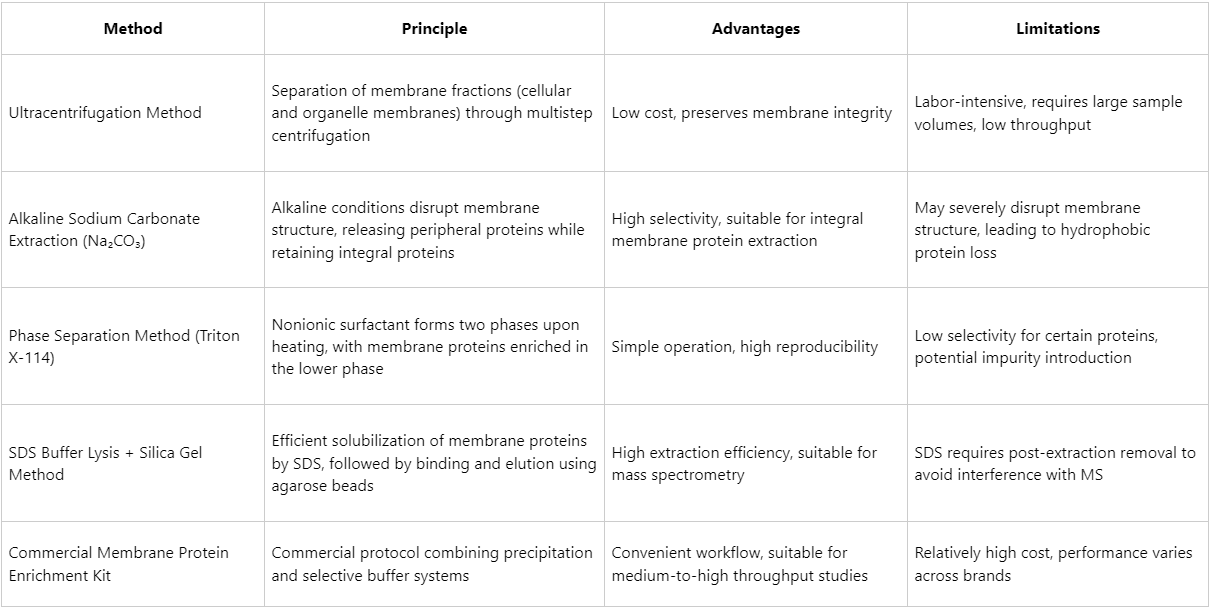
Recommended Strategies for Efficient Membrane Protein Enrichment
1. Differential Ultracentrifugation Combined with Density Gradient Purification
(1) Applicability: Ideal for large-scale samples (>10^8 cells) and studies requiring preservation of intact membrane structures.
(2) Workflow:
(3) Advantages: Enables subcellular fractionation and enrichment of membrane-associated proteins.
(4) Limitations: Requires access to an ultracentrifuge and involves prolonged processing time.
MtoZ Biolabs provides integrated workflows for the simultaneous enrichment of membrane and organelle proteins (e.g., mitochondria, endoplasmic reticulum), balancing structural specificity with high throughput.
2. Triton X-114 Phase Partitioning (Classical Yet Robust Approach)
(1) Applicability: Suitable for quantitative analysis of membrane proteins in cellular or tissue samples.
(2) Workflow:
(3) Advantages: Simple, reproducible, and amenable to parallel sample processing.
(4) Limitations: Extraction efficiency for peripheral membrane proteins may be suboptimal.
3. Combined Alkaline Carbonate Treatment and SDS Lysis
(1) Applicability: Designed for in-depth coverage of integral membrane proteins.
(2) Workflow:
(3) Advantages: Provides broad proteome coverage and is particularly compatible with mass spectrometry.
(4) Cautions: Complete removal of SDS is essential; desalting and optimized digestion protocols are recommended.
Post-Enrichment Mass Spectrometry and Data Analysis Recommendations
While enrichment enhances detection, subsequent steps in mass spectrometry adaptation and bioinformatic analysis are equally critical. Recommended best practices include:
MtoZ Biolabs’s Comprehensive Membrane Protein Enrichment and Proteomics Platform
MtoZ Biolabs has developed an integrated platform for customized membrane protein enrichment and mass spectrometric analysis, featuring:
Membrane protein research represents a systematic endeavor rather than a simple extraction–detection–identification workflow. Each stage, from sample preparation and buffer optimization to digestion strategies and instrument compatibility, dictates the depth and precision of analysis. MtoZ Biolabs is committed to addressing the challenges of membrane protein enrichment, extraction, and identification, offering tailored technical evaluations and consultations for researchers.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







