How to Choose the Appropriate Strategies for Ubiquitinated Protein Enrichment?
-
Broad applicability, capable of recognizing multiple ubiquitin chain types
-
Straightforward procedure, compatible with most standard laboratory workflows
-
Lack of chain-type specificity, as antibodies can recognize multiple ubiquitin linkages
-
Potential co-purification of a substantial number of non-target proteins, resulting in a complex mass spectrometry background
-
High binding affinity, markedly improving capture efficiency
-
Ability to selectively enrich specific ubiquitin chain types (e.g., K48 or K63)
-
Relatively complex design and higher cost
-
Requires a priori knowledge of the target chain type, making it less suitable for broad, exploratory applications
-
Precise localization of ubiquitination sites
-
Simplified downstream mass spectrometry analysis with a high signal-to-noise ratio
-
Applicable only to trypsin-digested samples
-
Limited coverage of peptides at the distal ends of ubiquitin chains or of small peptides
-
Time-intensive and associated with high antibody costs
-
Enables the simultaneous analysis of dozens of samples in a single mass spectrometry experiment
-
Generates large datasets with high quantitative accuracy
-
High cost of TMT reagents and increased complexity of experimental design
-
Requires advanced high-resolution mass spectrometers, such as Orbitrap Fusion Lumos
-
Clearly define research objectives: protein-level or site-level analysis? Chain-type-specific or broad-spectrum capture?
-
Evaluate budgetary constraints and available resources: antibody-based approaches offer moderate costs; isobaric labeling is suitable for large-scale, high-throughput studies.
-
Match the method to sample type and amount: high-affinity TUBEs for small samples, site-specific enrichment for complex samples.
-
Prioritize data quality: for projects requiring repeated acquisition of high-coverage, high-accuracy ubiquitination site data, collaboration with specialized service providers is recommended.
Ubiquitination, a key reversible post-translational modification of proteins, plays a central role in various biological processes, including cell cycle regulation, protein degradation, DNA damage repair, and signal transduction. The comprehensiveness and precision of ubiquitination research are often contingent upon the use of efficient and highly selective ubiquitinated protein enrichment strategies. Requirements for these strategies vary considerably depending on the experimental objectives, sample sources, and depth of analysis. Therefore, selecting an appropriate approach for ubiquitinated protein enrichment is a critical consideration at the experimental design stage. A well-chosen strategy not only impacts data quality but also directly influences research efficiency and the reliability of conclusions.
Overview of Enrichment Strategies
1. General Anti-Ubiquitin Antibody Enrichment
(1) Principle and procedure: Monoclonal ubiquitin antibodies (e.g., FK2, VU-1, or P4D1) are used to capture ubiquitin-modified proteins, enabling enrichment at the total proteome level.
(2) Advantages:
(3) Limitations:
(4) Application scenarios: Particularly suitable for global quantification of ubiquitinated proteins, exploratory studies, and preliminary screening of candidate proteins.
2. Tandem Ubiquitin Binding Entities (TUBEs)
(1) Principle and procedure: Tandemly arranged ubiquitin-binding domains with di- or multivalency increase affinity for ubiquitin chains, typically enabling capture of K48 and K63 linkages.
(2) Advantages:
(3) Limitations:
(4) Application scenarios: Well-suited to studies of polyubiquitin chains, such as investigations of protein degradation pathways and signal transduction mechanisms.
3. Site-Specific K-ε-GG Remnant Enrichment
(1) Principle and procedure: Following trypsin digestion of cell lysates, peptides containing Gly–Gly (K-ε-GG) remnants from ubiquitination are captured using antibodies that specifically recognize the modified lysine residue. This method directly enriches ubiquitination sites.
(2) Advantages:
(3) Limitations:
(4) Application scenarios: Ideal for ubiquitin site proteomics, quantitative comparisons, and detailed characterization of modification sites.
4. High-Throughput Peptide Capture with Isobaric Labeling (DiGly + TMT or TMTpro)
(1) Principle and procedure: Combines K-ε-GG site enrichment with TMT/TMTpro isobaric labeling to enable high-throughput, multiplexed comparison of multiple samples.
(2) Advantages:
(3) Limitations:
(4) Application scenarios: Particularly advantageous for dynamic ubiquitome profiling, drug intervention studies, and time-course experiments.
Key Parameters Influencing Strategy Selection
1. Research Objective: Protein-Level vs. Site-Specific Analysis
(1) For generating a comprehensive list of ubiquitin-modified proteins → Anti-ubiquitin antibody approach or TUBEs are generally adequate
(2) For precise mapping of ubiquitination sites → K-ε-GG enrichment is the preferred choice
2. Ubiquitin Chain-Type and Linkage Specificity
(1) K48-linked polyubiquitin chains: primarily associated with protein degradation
(2) K63-linked chains: involved in signal transduction and DNA repair
TUBEs can be selected based on chain type to effectively enrich proteins carrying the desired linkage.
3. Quantitative Requirements and Sample Throughput
(1) Studies with a small number of comparative groups (e.g., 2–4): label-free quantification (LFQ) with K-ε-GG enrichment is recommended
(2) Studies with multiple groups (≥5): TMT/TMTpro labeling combined with K-ε-GG enrichment and high-throughput LC-MS/MS is recommended
4. Sample Type and Starting Material
(1) Cell lines or small animal tissues (≤1 mg protein): TUBEs or antibody-based methods are generally adequate
(2) Large tissue volumes (≥10 mg protein): site-specific enrichment is advised to minimize non-specific capture, with K-ε-GG enrichment being the preferred option
5. Laboratory Resources and Budget Considerations
(1) Antibody-based and TUBEs approaches generally involve moderate costs, with antibody purchase being the primary expense
(2) K-ε-GG antibodies combined with TMT labeling reagents are cost-intensive, but yield high-quality site-specific data with significant long-term benefits
(3) Specialized service providers, MtoZ Biolabs, can supply small-scale reagents and customized protocols, helping to reduce R&D expenditure
Cost-Effectiveness Summary
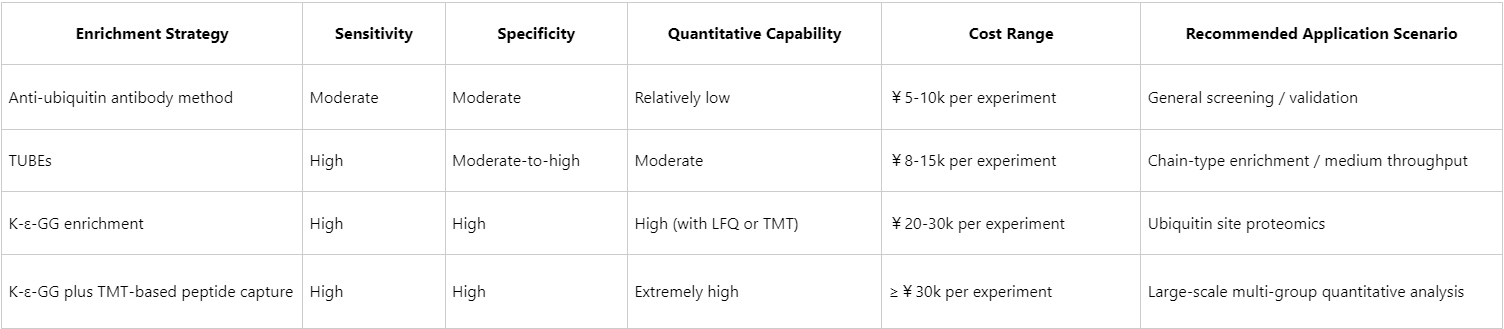
Note: Cost estimates are based on typical experiments; actual values depend on sample starting material, antibody/reagent brands, and experimental scale.
Example Workflow: K-ε-GG Enrichment
1. Sample Preparation
(1) Rapid lysis using RIPA or SDS-based buffer
(2) Protein quantification, preferably using the BCA or Bradford method
2. Protein Digestion
(1) Process: reduction → alkylation → trypsin digestion (4–16 h incubation)
(2) Optional: secondary digestion (LysC + trypsin) to enhance coverage
3. Pre-Enrichment Cleanup
Ion exchange (SCX/SAX) or high-pH reverse phase chromatography to remove interfering substances
4. K-ε-GG Antibody Enrichment
(1) Perform under cold-chain conditions; incubate at 4 °C for ≥1 h
(2) Remove unbound material; collect peptides after elution
5. Mass Spectrometry Analysis
(1) Nano-scale LC-MS/MS using high-resolution instruments such as Orbitrap Exploris or Fusion Lumos
(2) Database search with a false discovery rate (FDR) ≤ 1% for both protein and site identifications
6. Data Analysis
(1) Software options include MaxQuant, Proteome Discoverer with PTMProfiler, and MSstats
(2) Conduct quantitative comparisons, Gene Ontology (GO) enrichment, and ubiquitination site interaction network analysis
Principles for Selecting the Appropriate Ubiquitinated Protein Enrichment Strategy
By carefully selecting the most appropriate ubiquitinated protein enrichment approach, researchers can substantially improve mass spectrometry coverage and facilitate the accurate identification of critical ubiquitination sites, thereby enhancing the robustness and impact of their studies. Feel free to contact MtoZ Biolabs at any time. We will combine your project background to provide customized experimental design, execution, and data analysis, to accelerate the realization of research results.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







