How to Achieve Quantitative Mapping of Protein Localization in Tissues?
-
P53 participates in DNA damage repair when localized to the nucleus, whereas cytoplasmic P53 is involved in the regulation of apoptosis.
-
Membrane association of AKT kinase is a prerequisite for activation of its downstream signaling pathways.
-
Subcellular fractionation: Cells are separated into nuclear, mitochondrial, cytoplasmic, membrane, and other fractions using differential centrifugation, density gradient centrifugation, or commercial fractionation kits.
-
Protein extraction and enzymatic digestion: Proteins from each fraction are individually extracted and digested with trypsin.
-
Quantitative labeling: Isobaric labeling strategies such as TMT or iTRAQ, as well as label-free quantification methods, may be applied.
-
LC-MS/MS analysis: High-resolution mass spectrometry is employed for protein identification and quantification.
-
Data analysis: Localization prediction and visualization are performed by integrating reference databases (e.g., GO cellular component annotations), principal component analysis (PCA), and machine learning-based approaches.
-
High-throughput capability with quantitative output.
-
Enables analysis of protein relocalization under different experimental conditions.
-
Integration with global protein expression profiles allows in-depth characterization of dynamic localization changes.
-
The purity of subcellular fractions directly affects localization accuracy.
-
Loss of difficult-to-extract proteins, particularly membrane-associated proteins, may occur.
-
Experimental procedures are relatively complex when applied to tissue samples.
-
Spatial transcriptomic sequencing and proteomic measurements are performed on identical or adjacent tissue sections.
-
Spatial registration algorithms are used to integrate transcriptomic and proteomic datasets.
-
Protein distribution patterns within tissues are inferred based on the spatial localization of corresponding transcripts.
-
Target proteins are genetically fused to enzymes such as APEX or BioID and expressed in living cells.
-
These enzymes catalyze covalent labeling of proteins in close proximity within a short time window.
-
Labeled proteins are subsequently identified by mass spectrometry, thereby defining the local subcellular environment of the target protein.
-
Nanometer-scale spatial resolution.
-
Potential applicability to dynamic or time-resolved studies.
-
Highly effective for investigating protein–protein interactions and local microenvironments.
-
Optimized subcellular fractionation strategies applicable to both cellular and tissue samples.
-
A high-sensitivity Orbitrap Fusion Lumos platform supporting 10-plex and 16-plex TMT-based analyses.
-
Comprehensive bioinformatics workflows for localization prediction and data visualization.
-
Customizable protein relocalization study designs compatible with experimental conditions such as drug treatments and genetic perturbations.
Quantitative mapping of protein localization represents a critical foundation for understanding protein function, regulatory mechanisms, and cellular behavior under pathological conditions. With recent advances in mass spectrometry (MS) technologies, it has become feasible to perform high-throughput and quantitative analyses of protein localization at the subcellular level.
Core Significance of Quantitative Mapping of Protein Localization
Cellular function critically depends on accurate protein localization and its spatiotemporal regulation. Many proteins exhibit multiple subcellular localizations (moonlighting functions), and their biological activities are closely linked to their distribution among cellular compartments such as the plasma membrane, mitochondria, nucleus, and cytoplasm. For example:
Accordingly, systematic and quantitative profiling of protein distributions across subcellular structures in cells or tissues can yield essential insights into protein function, signaling pathway mechanisms, and potential disease-associated targets.
Mainstream Technical Approaches for Quantitative Mapping of Protein Localization
1. Subcellular Fractionation Coupled with Quantitative Mass Spectrometry
This approach represents one of the most widely adopted strategies.
(1) Overview of the Experimental Workflow
(2) Advantages
(3) Limitations
2. Integration of Proteomics with Spatial Transcriptomics
Spatial transcriptomics provides spatially resolved gene expression information in tissue sections. When integrated with proteomic data, it enables indirect inference of protein localization patterns.
(1) Technical Framework
(2) Application Potential
This strategy is particularly suitable for reconstructing region-specific protein expression maps at the tissue-section level and holds significant promise for studies of tumor heterogeneity and tissue development.
3. Biotinylation-Based Proximity Labeling Technologies (e.g., APEX, BioID)
(1) Principle
(2) Key Characteristics
Note: This approach relies on gene editing or overexpression systems and is primarily suited for mechanistic investigations rather than high-throughput proteomic profiling.
Key Considerations in Data Analysis and Localization Prediction
1. Multidimensional Data Integration
Protein abundance, peptide specificity, GO annotations, and curated subcellular marker protein databases should be jointly integrated to enhance the accuracy of localization predictions.
2. Visualization Tools
Dimensionality reduction techniques such as t-SNE and UMAP are commonly used for visualizing subcellular distribution patterns.
Bioinformatics tools and R packages (e.g., pRoloc and MSnbase) enable predictive modeling of subcellular localization and interactive data exploration.
3. Analysis of Protein Relocalization
In studies involving drug treatments or disease models, particular emphasis should be placed on detecting protein translocation events across subcellular compartments under different conditions.
MtoZ Biolabs offers multi-condition TMT-based experimental designs to enable precise quantification of such dynamic localization changes.
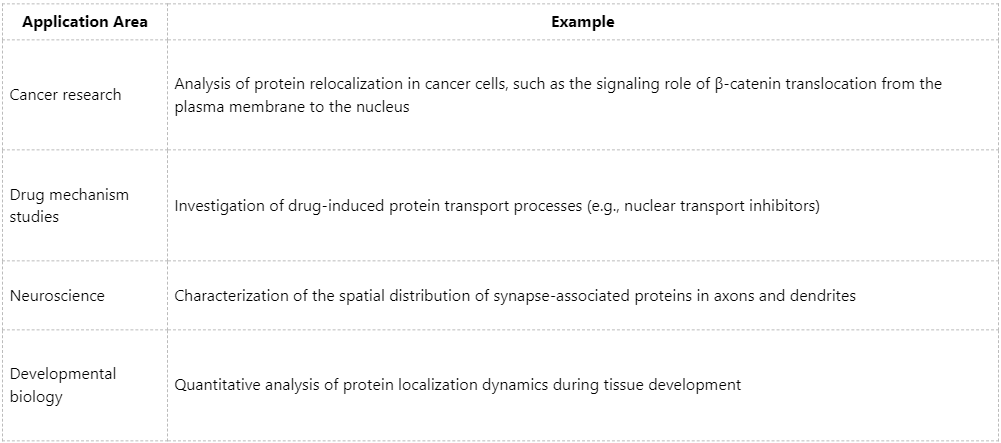
Application Scenarios and Research Value
Advantages of MtoZ Biolabs' Quantitative Protein Localization Solutions
For quantitative studies of subcellular protein localization, MtoZ Biolabs provides integrated high-resolution proteomics services, including:
Quantitative mapping of protein localization is emerging as a fundamental pillar of precision life science research. By integrating subcellular fractionation, quantitative mass spectrometry, and bioinformatics modeling, researchers can achieve unprecedented insights into the spatial dynamics of proteins. For studies focused on protein localization, MtoZ Biolabs provides end-to-end solutions ranging from sample preparation to advanced data interpretation, supporting high-impact research outcomes and publication.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







