How to Achieve High-Efficiency Protein Sequence Analysis with Edman Sequencing? A Lab Guide
-
Verification of N-terminal sequences in recombinant proteins or monoclonal antibodies
-
Characterization of protein truncations or post-translational modifications
-
Validation of sequences predicted by mass spectrometry
-
Analysis of samples with poor ionization efficiency or low mass spectrometric response
-
An automated Edman sequencing system
-
A standardized PVDF membrane transfer and sample pretreatment workflow
-
A validation protocol integrated with high-resolution mass spectrometry
-
Comprehensive technical documentation and detailed data reports to support research project applications
Edman sequencing is a classical and highly specific technique for protein N-terminal sequence analysis, which continues to play a pivotal role in structural proteomics, antibody engineering, and biopharmaceutical research. Although high-resolution mass spectrometry technologies have advanced rapidly, Edman sequencing retains unique, irreplaceable value in specific applications due to its ability to sequentially resolve N-terminal amino acids. This article provides a comprehensive laboratory guide on performing Edman degradation efficiently, covering the underlying principles, experimental procedures, sample preparation, data interpretation, and common troubleshooting strategies to support researchers in obtaining reliable protein sequence information.
What Is Edman Sequencing?
First introduced by Pehr Edman in 1950, Edman degradation is a chemical method that sequentially cleaves and identifies amino acid residues from the N-terminus of a polypeptide through cyclic reactions. The core mechanism involves the reaction of phenyl isothiocyanate (PITC) with the N-terminal amino acid to generate a stable derivative that can be selectively cleaved and analyzed, thereby enabling stepwise elucidation of the peptide sequence. In contrast to mass spectrometry, which is based on mass-to-charge ratio (m/z), Edman sequencing relies on chemical specificity, making it particularly suitable for verifying N-terminal integrity, detecting post-translational modifications or mutations, especially when the target protein is known.
Applications of Edman Sequencing
While modern proteomic analysis predominantly relies on mass spectrometry, Edman degradation remains advantageous in several specific contexts:
At MtoZ Biolabs, Edman degradation is routinely integrated with mass spectrometry-based methods to enable cross-validation of protein sequences, thereby enhancing the overall confidence and reliability of sequencing results.
Basic Procedure of Edman Sequencing
1. Sample Immobilization
For Edman degradation, protein samples must be immobilized on polyvinylidene difluoride (PVDF) membranes or glass fiber sheets to prevent loss of protein material during the reaction process. This immobilization is typically achieved through electrotransfer following SDS-PAGE. It is essential to avoid reagents that may interfere with the reaction, such as Tween-20.
2. N-Terminal Amino Acid Derivatization
The N-terminal amino group of the immobilized protein reacts with phenyl isothiocyanate (PITC) under alkaline conditions, forming a cyclic phenylthiohydantoin (PTH) derivative of the amino acid. As the reaction is highly sensitive to moisture, strict environmental control is required to maintain reaction fidelity.
3. Cleavage and Recovery
An anhydrous acid is subsequently introduced to cleave the peptide bond at the N-terminus, releasing the PTH-amino acid derivative. Efficient recovery of the PTH derivatives at this stage is crucial to ensure adequate analytical sensitivity in downstream identification.
4. Analysis and Identification
The collected PTH derivatives are analyzed using high-performance liquid chromatography (HPLC) or capillary electrophoresis. Identification of the N-terminal amino acid sequence is achieved by comparing the retention times of the PTH derivatives with those of standard amino acid derivatives.
Key Factors for Experimental Success
To obtain high-resolution sequence data using Edman sequencing, the following considerations are critical:
1. Sample Purity
Edman degradation is highly sensitive to sample impurities. The presence of contaminating proteins can generate overlapping or ambiguous signals, complicating the accurate interpretation of amino acid sequences.
2. N-Terminal Accessibility
Edman sequencing requires a free, unmodified N-terminus. If the N-terminal residue is chemically blocked (e.g., through acetylation, post-translational modification, or retention of the signal peptide), sequencing cannot proceed. Enzymatic cleavage or chemical treatment to remove such modifications may be necessary during sample preparation.
3. Transfer Integrity
Protein degradation or inefficient membrane transfer during or after electrophoresis can impair the overall yield and reaction efficiency of Edman degradation. Ensuring complete and intact transfer of proteins to the membrane is therefore essential.
4. Reaction Efficiency per Cycle
The efficiency of Edman degradation diminishes progressively with each successive reaction cycle. As a result, the method is typically most reliable for identifying the first 10 to 20 amino acids at the N-terminus of the protein.
Edman Sequencing vs. Mass Spectrometry: How to Choose?
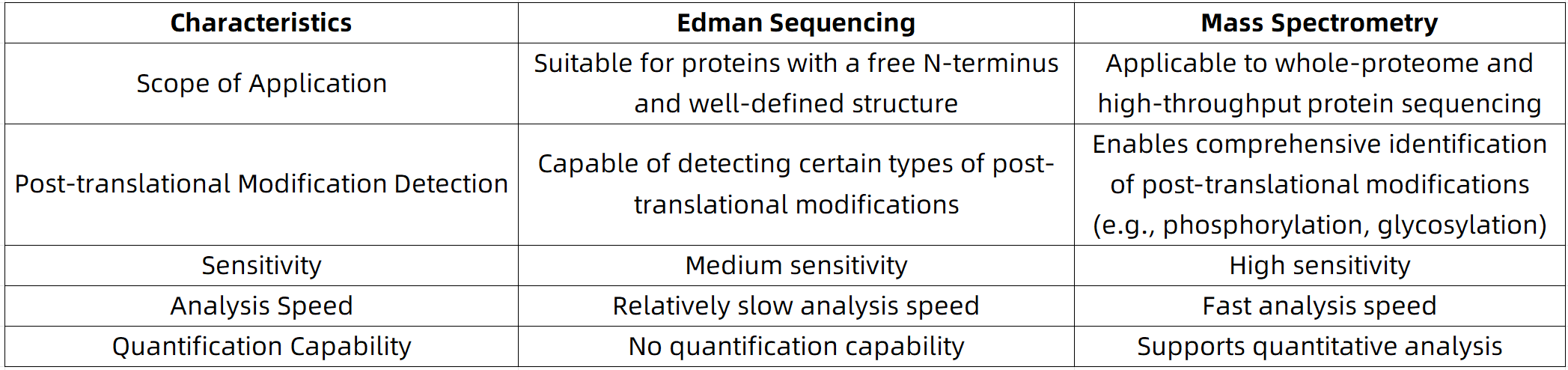
Suggested Strategy: When the integrity or modification of the N-terminal sequence is of primary concern, Edman sequencing remains the preferred method. For full-sequence acquisition or high-throughput analysis, mass spectrometry offers superior advantages. At MtoZ Biolabs, we offer an integrated protein sequencing solution that combines Edman degradation with high-resolution mass spectrometry, delivering a strategy tailored to each client’s specific needs.
At MtoZ Biolabs, we offer:
Whether your goals involve the quality assessment of monoclonal antibodies, detection of N-terminal modifications, or the generation of quality data for regulatory drug submissions, we provide protein sequencing analysis services that are both highly sensitive and highly reliable.
Although Edman sequencing is a well-established technique, it continues to hold distinct value in the analysis of protein N-terminal sequences. In applications such as structural verification, quality control in biopharmaceuticals, and protein engineering, accurate N-terminal information often plays a critical role. If you are seeking a protein sequence analysis service that is efficient, precise, and verifiable, MtoZ Biolabs is your trusted partner. We offer not only advanced analytical technologies but also reliable data support and collaborative research partnerships.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







