How PRM Proteomics Supports Biomarker Validation?
-
In the first-stage MS (MS1), the target peptide (precursor ion) is selected and introduced into the collision cell to generate multiple fragment ions.
-
The second-stage MS (MS2) performs a full scan to record the signals of all fragment ions.
-
Quantification and identification are achieved by matching the characteristic fragment ion spectra of the peptide.
-
Normalization using SIS peptides
-
Total area normalization
-
Median normalization across runs
-
Integrated experimental solutions from DDA-based discovery to PRM-based verification
-
A proprietary PRM template library covering tens of thousands of validated protein targets
-
Expert internal standard design and synthesis services for absolute quantification
-
Standardized analysis workflows (Skyline + R) delivering publication-quality figures and statistical outputs
-
Compatibility with diverse sample types, including serum/plasma, FFPE tissue, and cell lysates
Biomarkers are essential tools for early disease diagnosis, prognosis assessment, and therapeutic monitoring, and have become a focus of intensive research in biomedicine. However, how can clinically meaningful biomarkers be screened and validated from among tens of thousands of proteins? Parallel Reaction Monitoring (PRM), a highly specific and high-throughput targeted protein quantification technique, has emerged as a pivotal approach for advancing biomarker research from candidate identification to mechanistic validation and clinical translation. This article systematically describes the mechanisms, technical advantages, experimental design principles, and analytical workflows of PRM proteomics in biomarker validation, elucidates its unique value in proteomics research, and aims to facilitate efficient and reliable biomarker validation by researchers.
From Discovery to Validation: The Biomarker Validation Pathway
Biomarker research typically proceeds through three main stages:
1. Discovery
Untargeted proteomics approaches, such as Data-Dependent Acquisition (DDA) or Data-Independent Acquisition (DIA), are used to identify candidate proteins showing differential expression between distinct experimental or clinical groups.
2. Verification
Targeted approaches, such as PRM, are applied to quantitatively verify these candidate proteins with high precision in larger cohorts.
3. Clinical Validation
High-throughput platforms such as ELISA or mass spectrometry–based peptide arrays are employed to confirm their specificity and sensitivity in clinical cohorts.
PRM serves as a crucial bridge between the discovery and clinical validation phases. It combines strong target specificity and high sensitivity with the capacity for multiplexed analysis, making it particularly suitable for medium-scale validation tasks involving tens to hundreds of candidate peptide targets.
Principles and Advantages of PRM Technology
PRM is a targeted quantitative analysis method developed for high-resolution, high-mass accuracy mass spectrometers (e.g., Thermo Orbitrap series). Its core principles are as follows:
Key Technical Advantages of PRM Proteomics
1. High Specificity
PRM acquires multiple high-resolution fragment ion peaks simultaneously, effectively distinguishing co-eluting background signals and greatly enhancing selectivity. This is particularly advantageous for detecting low-abundance proteins in complex matrices such as plasma or tissue samples.
2. High Precision and Reproducibility
Unlike SRM/MRM, PRM does not require predefined ion pairs, allowing for broader acquisition coverage and greater flexibility in method development. Inter-assay reproducibility (coefficient of variation, CV) is typically maintained within 10%.
3. Traceability and Expandability
All acquired MS2 spectra can be re-mined for additional peptides post-acquisition, enabling target expansion or correction of prior analyses. This makes PRM well suited for iterative studies and multi-center collaborations.
4. Efficient Method Development
Target peptide lists can be generated directly from existing DDA/DIA datasets, and analysis methods can be rapidly built using software such as Skyline, reducing both development time and labor requirements.
Key Considerations in PRM Proteomics Experimental Design
Effective biomarker validation requires well-structured PRM experimental design. Key elements include:
1. Target Peptide Selection
(1) Peptides should be uniquely assigned to the target protein, with lengths generally between 7–20 amino acids.
(2) Avoid modification-prone sites, such as methionine oxidation or glutamine deamidation, whenever possible.
(3) Prioritize peptides with robust signal intensity in prior DDA/DIA experiments.
2. Internal Standard Design
(1) Stable isotope-labeled (SIS) peptides are recommended for achieving absolute quantification or improving relative quantification accuracy.
(2) Internal standards should match the retention time and approximate ionization efficiency of the endogenous peptides.
3. Instrument Parameter Optimization
(1) Employ high-resolution, high-mass accuracy Orbitrap instruments, with MS2 resolution set to 30,000 or higher.
(2) Define appropriate retention time windows and target fragment ion m/z values to maximize acquisition efficiency.
4. Biological Replicates and Quality Control
(1) Include at least three biological replicates for each sample group.
(2) Incorporate QC samples (e.g., pooled samples or SIS peptides) to monitor system stability throughout the experiment.
PRM Proteomics Data Analysis Workflow
1. Data Import and Target Definition
Raw mass spectrometry data are imputted into software such as Skyline, along with a target peptide list and corresponding fragment ions.
2. Chromatographic Peak Extraction and Identification
Extracted ion chromatograms (XICs) are generated based on predefined retention time windows and ion transitions, and chromatographic peaks are identified either automatically or manually.
3. Peak Integration and Quantification
Relative or absolute quantification is performed by integrating the area under selected fragment ion peaks, typically using three to five ions with high signal-to-noise ratios and strong reproducibility.
4. Data Normalization
Common approaches include:
5. Statistical Analysis
Processed quantitative data are subjected to statistical testing (e.g., t-test, Mann–Whitney U test), and diagnostic performance is evaluated using ROC curve metrics, including the area under the curve (AUC), sensitivity, and specificity.
6. Visualization and Reporting
Differential peptide information is visualized through volcano plots, heatmaps, and hierarchical clustering. Standardized reports are generated for project documentation and publication.
Comprehensive Advantages of PRM Proteomics in Biomarker Validation
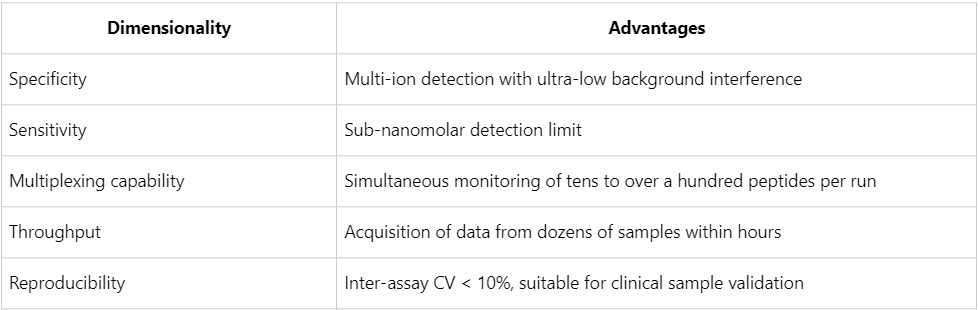
Leveraging the Orbitrap Fusion Lumos/Exploris platform, together with a high-coverage target database and an internally optimized PRM workflow, MtoZ Biolabs offers:
With its exceptional specificity, sensitivity, and reproducibility, PRM proteomics is regarded as an established reference standard for biomarker validation. Through rigorous experimental design, precise internal standard implementation, and standardized data analysis, researchers can efficiently and reliably identify protein biomarkers with strong translational potential from among numerous candidates. MtoZ Biolabs continues to refine its PRM mass spectrometry platform and data processing pipelines, thereby accelerating the translation of biomarkers from laboratory research to clinical practice and contributing to the advancement of precision medicine. Researchers may contact the company for project evaluation and methodological consultation as needed.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







