Exosome Protein Expression Verification Service
Exosome protein expression verification is a specialized technical service for qualitative and quantitative analysis of characteristic proteins in exosomes, aiming to confirm the origin, purity, and biological function of exosomes. This service is based on classical protein detection methods such as Western blot, ELISA, and mass spectrometry (MS). By identifying specific protein markers on the surface or inside exosomes (such as CD63, CD81, TSG101), it verifies the purity of exosomes in the sample and their protein expression profiles, thereby clarifying their biological function or origin.
Exosome protein expression verification service is widely applied in areas such as cancer research, drug delivery, immune regulation, and neurodegenerative diseases. It is particularly suitable for the development of exosomes as potential biomarkers and for preclinical functional validation. Accurate verification of protein expression in exosomes allows for in-depth investigation of their mechanisms in disease progression and supports the design and clinical translation of exosome-based therapeutic strategies.
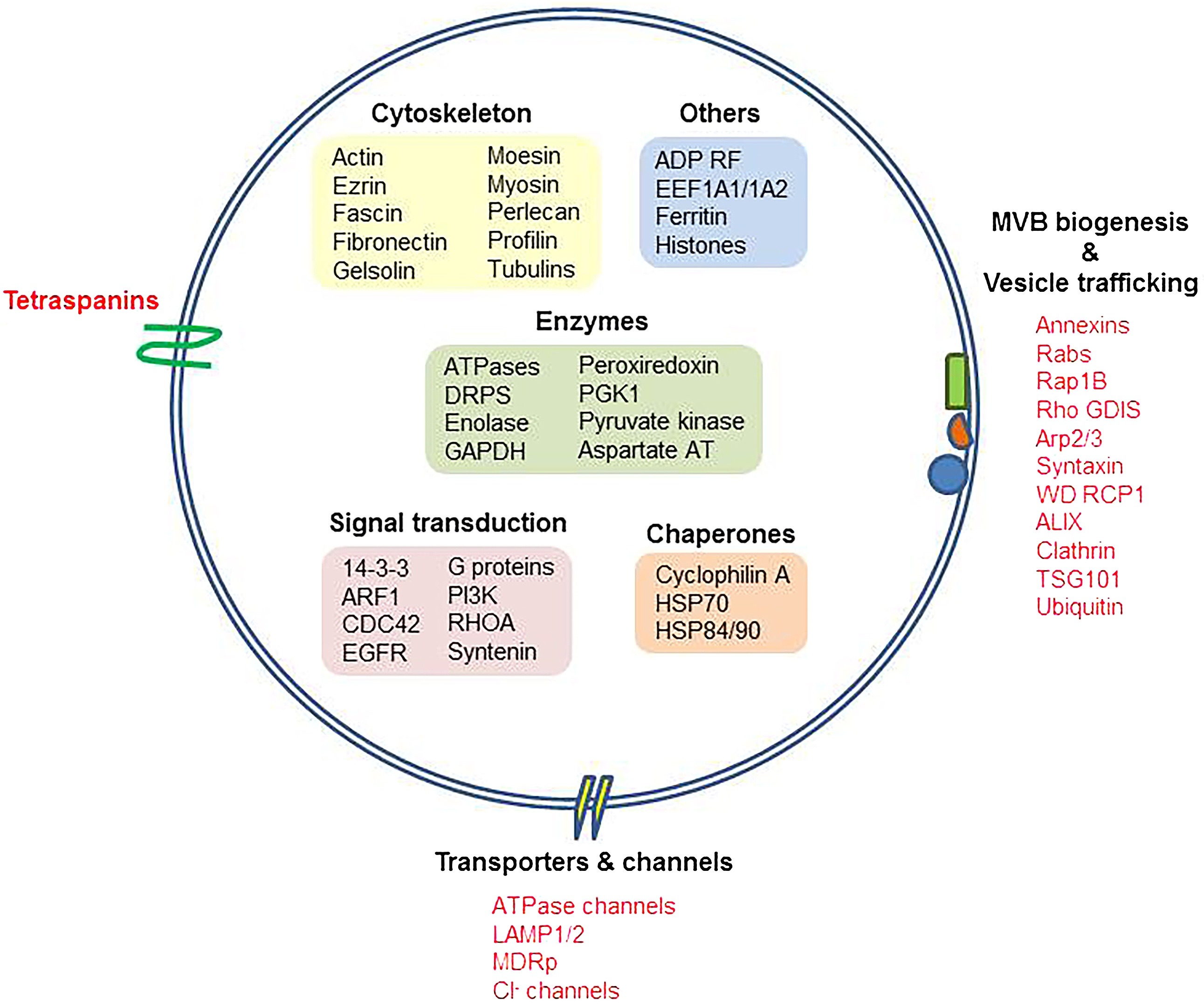
Siles-Lucas, M. et al. Veterinary Parasitology, 2017.
Figure 1. Exosome Signatures and Exosomal Proteins.
Services at MtoZ Biolabs
Based on advanced platforms such as Western blot, enzyme-linked immunosorbent assay (ELISA), flow cytometry (FACS), and high-resolution mass spectrometry (LC-MS/MS), the exosome protein expression verification service provided by MtoZ Biolabs enables qualitative and quantitative analysis of key proteins on the surface and inside exosomes (such as CD63, CD81, Alix, TSG101). The service includes exosome protein extraction, expression verification, and data analysis, ensuring high sensitivity, high specificity, quantitative accuracy, and reproducibility. This strongly supports research and development related to exosome functions and biomarker discovery.
Service Advantages
1. Multi-platform Integrated Analysis
Utilizes advanced instruments including Western blot, ELISA, flow cytometry (FACS), and high-resolution mass spectrometry (LC-MS/MS) to ensure data accuracy.
2. Accurate Quantification
The service offers high sensitivity and specificity, enabling precise quantification of exosome-specific protein markers.
3. Standardized Workflow
Establishes a well-defined protocol for protein extraction and analysis to enhance experimental consistency and data comparability.
4. Customized Solutions
Offers flexible analysis strategies and technical support tailored to different research objectives and sample types, meeting the needs of both basic research and translational applications.
Applications
1. Exosome Purity and Origin Verification
By detecting specific protein markers, this service verifies the purity and origin of isolated exosome samples, ensuring the reliability of downstream experiments.
2. Disease Biomarker Research
The exosome protein expression verification service can be used to identify differential expression of exosomal proteins associated with specific diseases, advancing biomarker development in fields such as oncology and neurodegenerative disorders.
3. Exosome Function Mechanism Exploration
By integrating protein expression profiles, this service helps analyze the roles of exosomes in biological processes such as cell communication, immune regulation, and inflammatory responses.
4. Quality Control for Drug Delivery Carriers
In gene therapy or nanomedicine development, the exosome protein expression verification service can be used to validate the expression of target proteins in engineered exosomes, supporting evaluation of delivery efficiency and safety.
Case Study
1. Lymphocyte Migration Regulation Related Proteins in Urine Exosomes May Serve as a Potential Biomarker for Lung Cancer Diagnosis
This study aimed to evaluate the potential of proteins related to lymphocyte migration regulation in urinary exosomes as non-invasive biomarkers for lung cancer diagnosis. The study involved urine samples collected from patients diagnosed with lung cancer and healthy controls. Exosomes were isolated from urine and analyzed using proteomic techniques to assess the expression levels of proteins associated with lymphocyte migration. The expression profiles were then compared between the lung cancer group and the healthy control group. The results revealed significant upregulation or downregulation of certain lymphocyte migration-regulating proteins in the urinary exosomes of lung cancer patients compared to healthy individuals. The study concluded that these proteins could serve as promising non-invasive biomarkers for lung cancer diagnosis, offering a new approach for early detection and diagnostic evaluation.
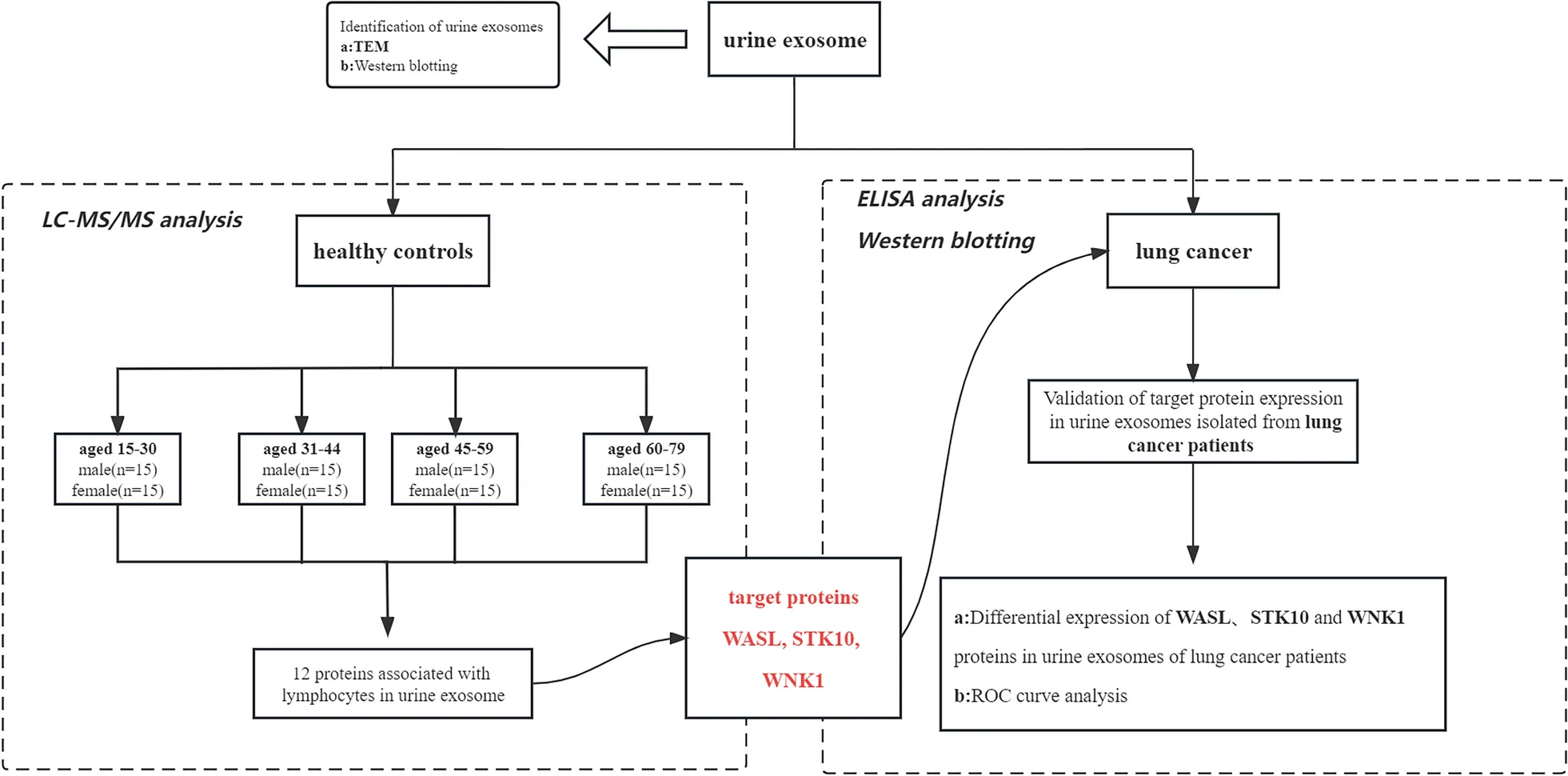
Jin, S. et al. BMC Cancer, 2023.
Figure 2. The Detailed Workflow of this Study.
2. Comprehensive Analysis and Comparison of Proteins in Salivary Exosomes of Climacteric and Adolescent Females
This study aimed to comprehensively analyze and compare the protein composition of salivary exosomes in climacteric and adolescent females to reveal the biological differences between these physiological stages. The study subjects included climacteric and adolescent women. Exosomes were isolated from the saliva of both groups, and high-resolution mass spectrometry was used to perform quantitative proteomic analysis. Bioinformatic tools were applied to annotate the differentially expressed proteins and conduct pathway enrichment analysis. The results showed that salivary exosomes from climacteric females exhibited differential expression of various proteins mainly involved in immune regulation, metabolic processes, and cell communication. The study concluded that changes in the salivary exosome proteome reflect stage-specific biological characteristics in females and may offer new insights into physiological transitions and disease risks associated with menopause.
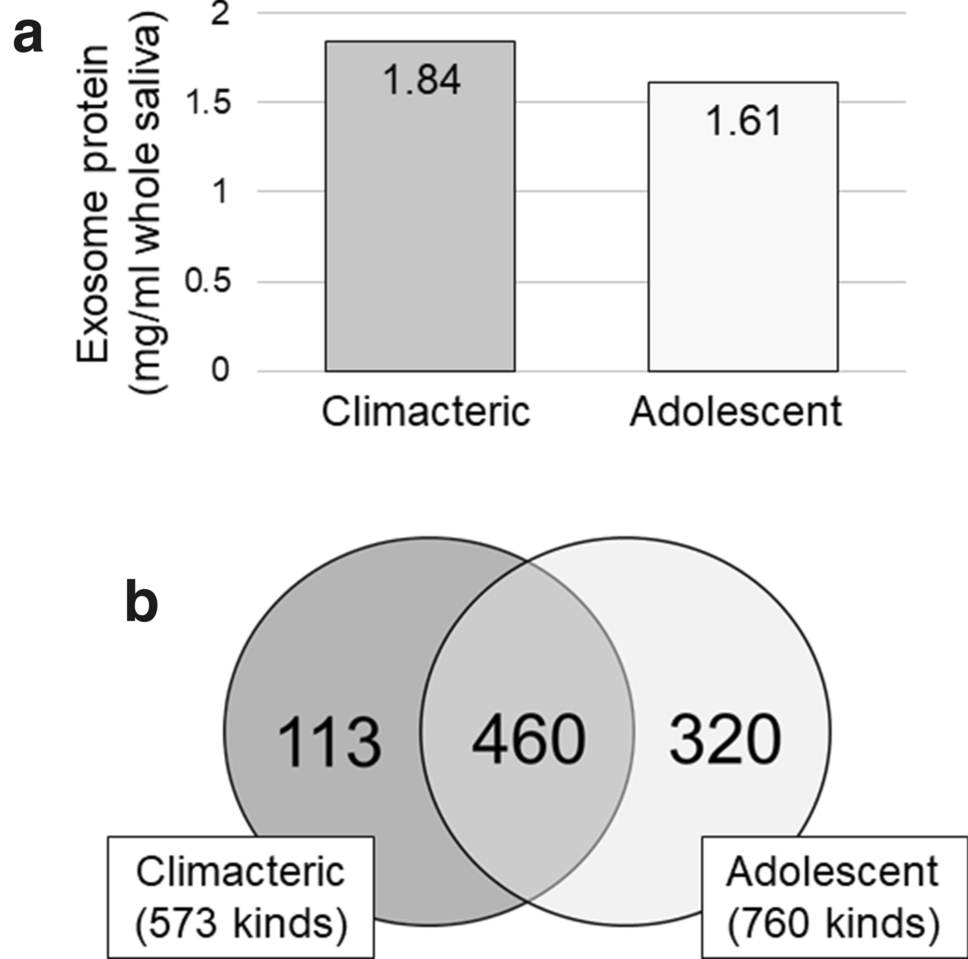
Lmai, A. et al. Odontology, 2021.
Figure 3. Total Exosome Protein in 1 mL Saliva (a) and Numbers of Protein Types (b) in Climacteric and Adolescent Exosomes.
FAQ
Q1: What Types of Samples Are Supported by the Service?
A1: The service supports exosomes derived from various sample types, including cell culture supernatant, serum, plasma, urine, cerebrospinal fluid, and breast milk. For other sample types, please feel free to contact us for detailed consultation.
Q2: Can Multiple Samples Be Analyzed Simultaneously?
A2: Yes. We offer high-throughput processing capabilities and can perform parallel detection and comparative analysis across multiple sample groups, making it suitable for studies such as differential expression screening.
How to order?







