Essential Analysis Tools for Label-Free Proteomics: A Comprehensive Review of MaxQuant and DIA-NN
Label-Free Proteomics has gained widespread application in basic biological research, disease mechanism elucidation, and biomarker discovery due to its streamlined workflow, relatively low cost, and broad applicability. However, the processing of LFQ data remains challenging, encompassing large-scale mass spectrometry data handling such as feature extraction, quantification, differential analysis, and functional annotation. Therefore, selecting and effectively utilizing appropriate analytical tools is critical for researchers. This review systematically examines two widely used tools for label-free proteomics—MaxQuant and DIA-NN—and provides an authoritative and practical reference for their application.
MaxQuant—A Benchmark in Classical Label-Free Proteomics
1. Introduction to MaxQuant
MaxQuant is a mass spectrometry data analysis tool developed by the Max Planck Institute in Germany. It is designed to support label-free quantification, SILAC, and TMT/iTRAQ workflows, and is widely used in proteomics research under the data-dependent acquisition (DDA) mode.
Key features include:
(1) Compatibility with high-resolution mass spectrometry platforms (e.g., Orbitrap)
(2) Integrated Label-Free Quantification (LFQ) algorithm
(3) Robust Match Between Runs (MBR) functionality that enhances data completeness across samples
(4) Seamless integration with Perseus for downstream statistical analysis and visualization
2. Core Functionalities of MaxQuant
(1) Feature Extraction and Identification
Utilizes the Andromeda search engine to perform efficient peptide and protein-level identification.
(2) Label-Free Quantification (LFQ)
Implements the MaxLFQ algorithm to normalize peptide intensities and enable relative quantification across experimental groups.
(3) Handling Missing Values and Match Between Runs (MBR)
Employs the MBR strategy to propagate peptide identifications across runs, improving overall proteome coverage.
(4) Quality Control and Visualization
Generates comprehensive quality control metrics (e.g., mass calibration error, peptide length distribution) and visualization outputs to assist in evaluating data quality.
DIA-NN—A Robust Engine for Processing Label-Free Proteomics DIA Data
1. Introduction to DIA-NN
DIA-NN (Data-Independent Acquisition Neural Networks) is an open-source software developed by the team led by Russian scientist Dmitry Kolmogorov. It is specifically designed for processing data generated by Data-Independent Acquisition (DIA) workflows. In recent years, DIA-NN has emerged as a widely adopted tool in label-free quantitative proteomics, particularly valued for its high-throughput capability and comprehensive proteome coverage.
Key Features:
(1) Utilizes deep neural networks for spectral prediction and protein quantification
(2) Supports library-free analysis workflows, eliminating the need for experimental spectral libraries
(3) Offers exceptionally fast data processing, enabling efficient handling of large datasets
(4) Achieves high sensitivity and quantitative accuracy, making it particularly suitable for large-scale sample analysis
2. Core Functionalities of DIA-NN
(1) Spectral Library Generation and Prediction
DIA-NN can generate theoretical spectral libraries directly from FASTA sequences, thus eliminating the traditional reliance on empirically derived spectral libraries.
(2) Protein Quantification and Data Normalization
The software integrates advanced normalization algorithms, including Cross-Run Normalization, to improve the comparability of quantitative results across different sample batches.
(3) Advanced FDR Control Strategies
DIA-NN applies hybrid approaches that incorporate neural network-based outputs to enhance the accuracy of identifying low-abundance proteins.
(4) Automated Batch Processing
It supports automated analysis workflows capable of processing hundreds to thousands of samples, making it well-suited for large-scale clinical cohort studies.
MaxQuant vs. DIA-NN: Selecting the Optimal Tool for Label-Free Proteomics Analysis
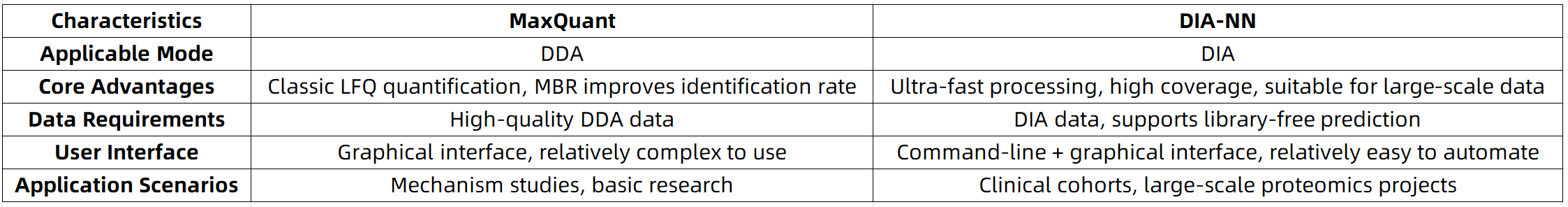
1. If your project is based on DDA data, involves a moderate number of samples, and prioritizes in-depth protein identification, MaxQuant is a well-suited tool;
2. If your study employs DIA data, requires high-throughput sample processing, or demands highly consistent quantification, DIA-NN is clearly the more suitable choice.
The success of label-free proteomics studies depends not only on high-quality mass spectrometry data, but also on the selection of appropriate data processing tools and specialized analytical strategies. MaxQuant has demonstrated robust performance under the conventional DDA acquisition mode, while DIA-NN represents a transformative advancement in DIA data analysis—together expanding the horizons of proteomics research. As a specialized provider of proteomics services, MtoZ Biolabs is dedicated to integrating globally advanced analytical platforms and standardized workflows to deliver tailored, reliable label-free quantification solutions for your research needs. Please contact us to learn more about the applications of MaxQuant and DIA-NN, as well as our high-quality proteomics service offerings.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







