Edman Sequencing vs. Mass Spectrometry: A Comparison of Protein Sequencing Techniques II
Proteins, as central functional molecules within cells, carry amino acid sequence information that not only elucidates their structure and function but also underpins various applications such as biomarker discovery, antibody characterization, and vaccine development. As a fundamental technique for deciphering amino acid composition and sequence, protein sequencing has evolved over several decades into two dominant methodologies: Edman sequencing and Mass Spectrometry (MS). What are the unique advantages of each of these protein sequencing approaches, and in what research contexts are they most effectively applied? This paper presents a comprehensive comparative analysis.
What Is Edman Sequencing?
Edman sequencing is a classical protein sequencing method developed by Pehr Edman in 1950. It is based on a stepwise chemical reaction that selectively cleaves and identifies amino acids from the N-terminus of a protein or peptide, thereby determining the sequence incrementally.
1. Core Mechanism
(1) PITC labeling: Phenyl isothiocyanate (PITC) reacts with the N-terminal amino acid to form a cyclic derivative;
(2) Acid cleavage: The labeled amino acid derivative, phenylthiohydantoin-amino acid (PTH-aa), is cleaved and released;
(3) HPLC analysis: The released PTH-aa is identified through high-performance liquid chromatography;
(4) The above steps are repeated iteratively to determine the amino acid sequence one residue at a time.
2. Advantages
(1) High sequencing fidelity, particularly suitable for validating known sequences or confirming antibody specificity;
(2) Effective for proteins with high purity and minimal contaminants;
(3) Relatively unaffected by certain amino acid side-chain modifications.
3. Limitations
(1) Typically limited to sequencing the first 30–50 residues from the N-terminus;
(2) Requires samples of high purity and relatively large amounts;
(3) Ineffective for sequencing internal or C-terminal regions of proteins;
(4) Incapable of detecting post-translational modifications (PTMs).
What Is Mass Spectrometry-based Protein Sequencing?
Mass spectrometry-based protein sequencing is a modern analytical technique that determines protein sequences by accurately measuring the mass-to-charge ratio (m/z) of peptide ions, in combination with database searches or de novo sequencing algorithms.
1. Core Process
(1) Proteolytic digestion: Proteins are enzymatically cleaved into smaller peptides, typically using trypsin;
(2) LC-MS/MS analysis: Peptides are first separated by liquid chromatography, followed by fragmentation and detection via tandem mass spectrometry;
(3) Sequence determination: Peptide spectra are interpreted through database matching or de novo computational inference.
2. Advantages
(1) High throughput: Enables simultaneous identification of thousands of proteins;
(2) High sensitivity: Capable of sequencing from samples at nanogram or even picogram levels;
(3) Post-translational modification (PTM) identification: Detects modifications such as phosphorylation, acetylation, and glycosylation;
(4) Applicability to complex biological samples: Suitable for analyzing cell lysates, tissue extracts, plasma, and other heterogeneous matrices.
3. Limitations
(1) Limited resolution for short peptides (< 5 amino acids);
(2) de novo sequencing results require experimental validation and may be subject to uncertainty;
(3) Identification challenges may arise from complex modifications and the presence of structural isomers.
Comparison Between Edman Sequencing and Mass Spectrometry
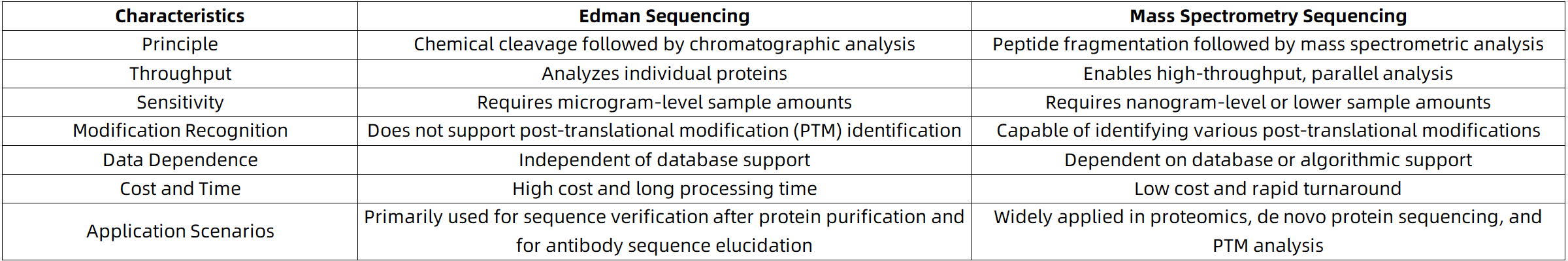
Suggestions for Choosing Protein Sequencing Techniques
If a highly purified single protein is available and the goal is to verify its N-terminal sequence or antibody fragment, Edman sequencing remains a valuable method.
Conversely, for comprehensive analysis of the proteome, post-translational modifications, or de novo sequencing of unknown proteins, mass spectrometry represents a more modern and efficient approach.
With the increasing demand for protein sequencing, selecting a reliable experimental platform has become critical. MtoZ Biolabs, equipped with advanced Orbitrap and Q-TOF mass spectrometry systems, offers a wide range of services, including N/C-terminal sequencing, de novo peptide sequencing, and post-translational modification analysis. These services are designed to meet the essential research needs of scientists and to support the biopharmaceutical industry in advancing antibody drug development, vaccine validation, and innovative protein design. Whether you are affiliated with a research institution, a hospital, or a biopharmaceutical company, MtoZ Biolabs provides a comprehensive solution for your protein sequencing challenges.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







