Edman Sequencing: Principles, Methods, and Key Technologies
-
N-terminal labeling: PITC reacts with the free amino group at the polypeptide N-terminus to form a phenylthiocarbamoyl peptide.
-
Cyclization and cleavage: Under acidic conditions, the N-terminal amino acid is selectively cleaved from the peptide chain, forming a cyclic PTH-amino acid, while the peptide chain is shortened by the removal of one residue.
-
PTH-amino acid analysis: The resulting PTH-amino acid is identified by high-performance liquid chromatography (HPLC) or other analytical methods, enabling deduction of the original protein's N-terminal amino acid sequence.
-
Cycle repetition: The above steps can be repeated iteratively, with one amino acid being cleaved and analyzed per cycle, ultimately allowing determination of the protein's N-terminal sequence.
-
It is applicable only to proteins with an unblocked N-terminus (e.g., proteins with N-terminal acetylation or methylation cannot be sequenced).
-
It is generally suitable for peptides up to 50 amino acids in length; beyond this, signal attenuation occurs due to decreased cycle efficiency.
-
It requires highly purified single protein samples and is not suitable for the direct analysis of complex mixtures.
-
Employing enzymatic or chemical treatments to remove modifications
-
Using mass spectrometry to predict the type and extent of modifications in advance
Edman degradation is a classical method for sequencing the N-terminus of proteins and is widely utilized in the analysis of primary protein structure. Although mass spectrometry has become the dominant technology in proteomics in recent years, Edman degradation retains unique advantages for the precise determination of N-terminal amino acid sequences. This paper introduces the basic principles and experimental methods of Edman degradation and discusses key strategies for its optimization.
Core Principles of Edman Degradation
Edman degradation, developed by the Swedish scientist Pehr Edman in 1950, is based on the selective cleavage of a single amino acid from the N-terminus of a polypeptide using phenyl isothiocyanate (PITC), followed by identification of the amino acid as a stable phenylthiohydantoin (PTH) derivative. The process comprises the following steps:
Edman degradation offers high specificity, enabling accurate determination of short peptide N-terminal sequences with minimal sample requirements. However, several limitations exist:
Methods and Experimental Procedures
Early implementations of Edman sequencing relied on manual procedures, which were time-consuming and inefficient due to their low throughput. Modern protein sequencers have greatly improved sequencing efficiency and reproducibility by automating the control of reaction parameters, such as temperature and the sequence of reagent addition, as well as integrating real-time online detection systems. Traditional high-performance liquid chromatography (HPLC) separates and identifies PTH-amino acids based on their retention times, while coupling with mass spectrometry enables more accurate identification through precise molecular weight analysis. This approach is particularly effective for detecting modified amino acids. The standard Edman degradation workflow consists of three critical stages: sample preparation, chemical reaction, and product detection.
1. Sample Preparation
The target protein must be a purified, homogeneous species to prevent signal interference from protein mixtures. Samples are typically immobilized on a solid-phase support, such as a PVDF membrane or glass fiber membrane, to enhance reaction efficiency. The length of the polypeptide chain is generally limited to 30–50 amino acids, as excessive sequencing cycles can result in signal degradation.
2. Chemical Reaction
Under alkaline conditions, phenylisothiocyanate (PITC) reacts with the α-amino group at the N-terminus of the protein. The resulting derivatized amino acid is cleaved from the polypeptide via acid hydrolysis. The liberated PTH-amino acid is then extracted with an organic solvent and subjected to analytical detection.
3. Product Detection and Sequence Interpretation
HPLC remains the most widely employed technique for detecting PTH-amino acids, owing to its high resolution and sensitivity. By comparing elution profiles with reference standards, the N-terminal amino acid sequence can be deduced step-by-step.
Key Factors Affecting the Performance of Edman Sequencing
1. Sample Purity and Homogeneity
Contaminants or peptide mixtures can produce overlapping signals, leading to reduced sequence readability.
2. N-Terminal Blockage or Modification
N-terminal modifications such as acetylation or formylation can prevent PITC from binding effectively. Strategies to address this include:
3. Membrane Immobilization Efficiency and Recovery
The protein sample must be securely immobilized on the solid-phase support (e.g., PVDF membrane) to minimize sample loss during the reaction process.
4. Side Reactions and Background Interference
Careful control of pH, temperature, and reaction time is essential to minimize PTH-amino acid degradation and to prevent failure in PTH-ring formation.
Optimization Strategies for Edman Sequencing: From Feasibility to Precision
At MtoZ Biolabs, we have significantly enhanced both the reliability and sequencing quality of Edman degradation through the following approaches:
1. Parallel multi-channel sample processing – Minimizes human-induced variability and increases throughput
2. High-sensitivity HPLC detection system – Enables reliable identification of PTH-amino acids at sub-nanomolar concentrations
3. Tailored protocols for N-terminal deprotection – Integrates enzymatic and chemical strategies to restore the native N-terminus
4. Mass spectrometry-assisted data integration – Correlates Edman-derived sequences with high-resolution MS data to ensure sequence consistency
5. Fully traceable SOP system – Developed in accordance with standards for drug development, suitable for regulatory submissions
Application Scenarios: Why Edman Sequencing Remains Essential
Despite its lower throughput compared to mass spectrometry, Edman degradation remains highly valuable in the following contexts:
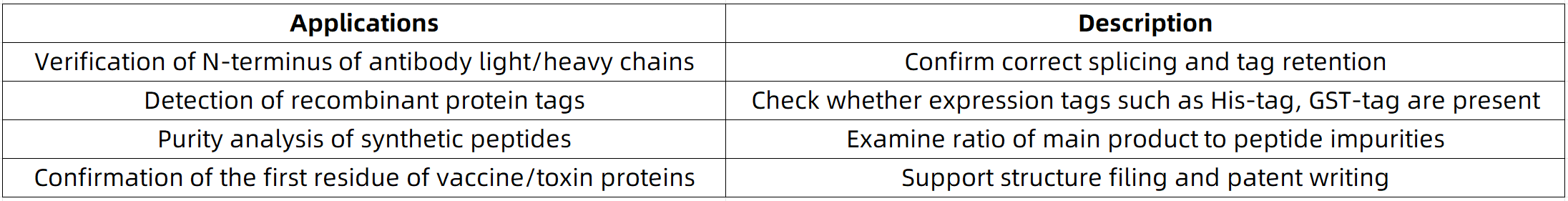
Edman sequencing remains indispensable in specific domains such as quality control of recombinant proteins (e.g., verifying N-terminal fidelity), biomarker discovery (e.g., detecting truncated peptides), and paleoproteomics (e.g., sequence characterization of degraded fossil peptides). While mass spectrometry dominates high-throughput proteomics, Edman degradation uniquely offers direct identification of N-terminal modifications (e.g., pyroglutamylation). MtoZ Biolabs, as a specialized provider of proteomics and mass spectrometry services, delivers high-precision N-terminal sequencing based on Edman chemistry.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







