Disulfide Bond Mapping Service
Disulfide Bond Mapping Service combines mass spectrometry and biochemical techniques to analyze the distribution and linkage of disulfide bonds in proteins. Disulfide bonds are crucial covalent bonds in proteins that play key roles in maintaining spatial conformation, regulating functional activity, and enhancing thermal stability. By revealing the disulfide bond connection patterns,Disulfide Bond Mapping Service enables researchers to gain a deeper understanding of protein higher-order structures and their biological functions. Leveraging advanced mass spectrometry platforms, MtoZ Biolabs offers Disulfide Bond Mapping Service including the qualitative, quantitative and positional analysis of disulfide bonds. This service can be applied to individual proteins as well as proteome-level studies, supporting protein structure elucidation and drug development efforts.
Analysis Workflow
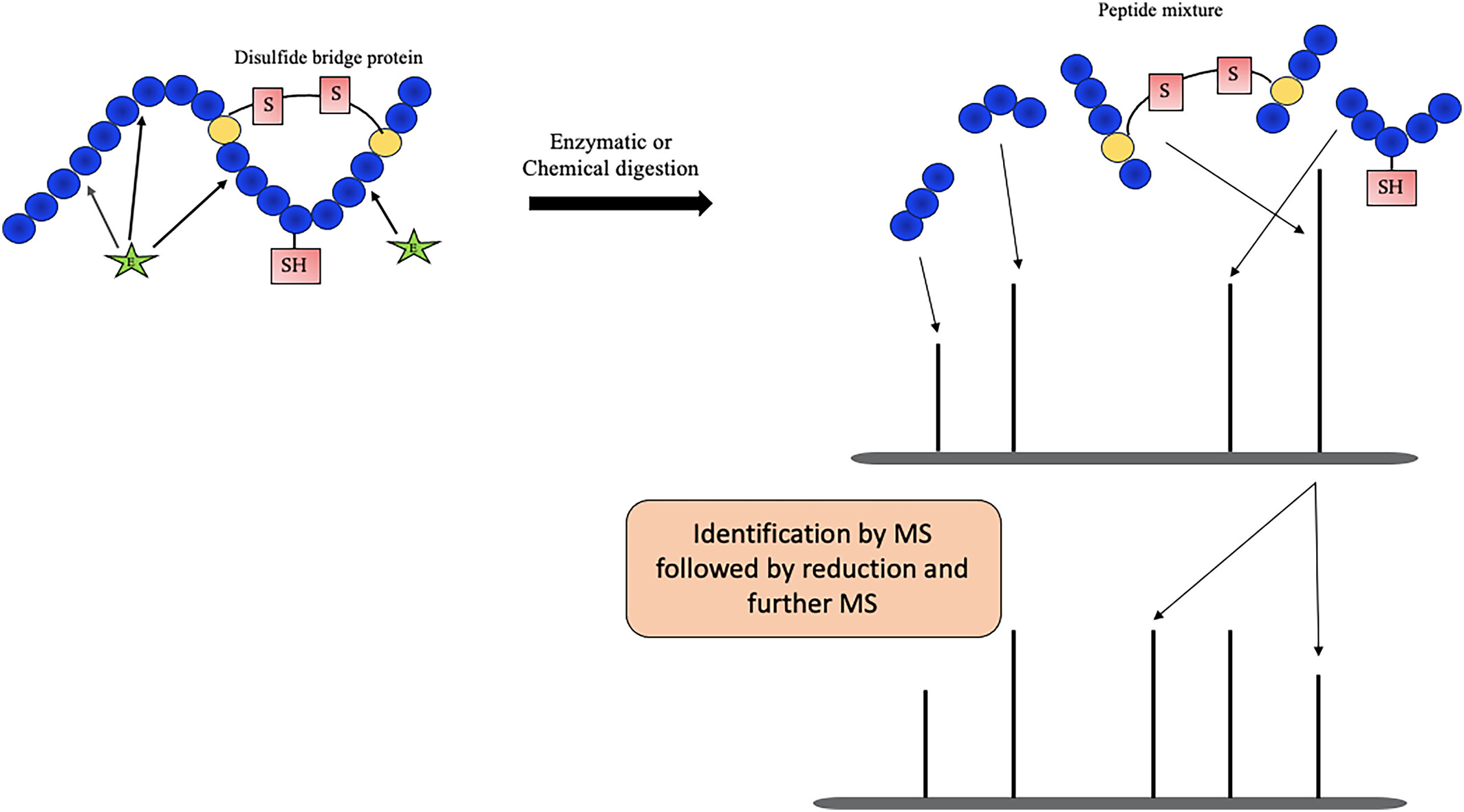
Chaturvedi S. et al. Rapid Commun Mass Sp. 2024.
General workflow for Disulfide Bond Mapping Service:
1. Sample Preparation
Extract the target protein and remove impurities.
Select appropriate enzymes for digestion based on the sample characteristics.
2. Non-Reducing Condition Analysis
Perform liquid chromatography separation and mass spectrometry analysis under non-reducing conditions to capture peptide information containing disulfide bonds.
3. Reducing Condition Verification
Use reducing agents such as DTT or TCEP to break disulfide bonds, followed by mass spectrometry analysis to verify changes in peptides after bond reduction.
4. Data Analysis and Mapping
Utilize specialized software and databases for data interpretation, accurately determining disulfide bond linkage sites and constructing protein disulfide bond maps.
5. Results Reporting
Deliver a comprehensive report, including peptide sequences, disulfide bond linkage patterns, and peptide distribution maps.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Disulfide Bond Mapping Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Disulfide Bond Mapping Service data, providing clients with a comprehensive data report.
Applications
Protein Structure Analysis
Determine the disulfide bond connection patterns of proteins to reveal their higher-order structure and stability.
Protein Function Studies
Investigate the role of disulfide bonds in protein folding, stability, and activity.
Antibody and Biopharmaceutical Development
Verify the correctness of disulfide bonds in antibody drugs and other protein therapeutics to ensure product consistency and stability.
Disease Mechanism Research
Analyze the impact of disulfide bond abnormalities (e.g., breakage or mispairing) on disease progression.
FAQ
Q. How to Improve the Coverage of Disulfide-Bonded Peptides in Disulfide Bond Mapping Experiments?
1. Select Suitable Digestion Strategies
Multi-Enzyme Digestion: Use a combination of proteases (e.g., trypsin, Glu-C, Lys-C) to increase peptide diversity and cover more disulfide-bonded regions.
Control Digestion Conditions: Optimize enzyme-to-substrate ratios, temperature, and incubation time to avoid overdigestion or incomplete digestion.
2. Optimize Peptide Separation
High-Performance Liquid Chromatography (HPLC): Use reverse-phase or nano-liquid chromatography (Nano-LC) under non-reducing conditions to finely separate complex peptides and reduce interference peaks.
Fractionation: Combine strong cation exchange (SCX) or high-pH reverse-phase fractionation to further separate complex samples, increasing the detection probability of low-abundance disulfide-bonded peptides.
3. Optimize Mass Spectrometry Parameters
Choose Appropriate Fragmentation Modes: Use collision-induced dissociation (CID), electron transfer dissociation (ETD), or hybrid modes like EThcD for better resolution of disulfide linkage sites.
Employ Data-Independent Acquisition (DIA): DIA provides broader coverage for low-abundance peptides, increasing peptide identification rates.
4. Enrich Target Peptides
Chemical Modifications or Antibody Capture: Enrich target peptide regions through chemical tagging or antibody-based capture to improve detection sensitivity.
High-Selectivity Enrichment Techniques: Use methods like IMAC or TiO₂ to simultaneously enrich for peptides with post-translational modifications and disulfide-bonded regions.
By implementing these strategies, the coverage of disulfide-bonded peptides in your samples can be significantly enhanced, ensuring more comprehensive and reliable experimental results.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This article used mass spectrometry to perform disulfide bond mapping and found that protein disulfide isomerase (PDI) can cleave specific disulfide bonds in histidine-rich glycoprotein (HRG), causing conformational changes in HRG and weakening its anti-thrombotic function, thereby promoting platelet activation and thrombosis.
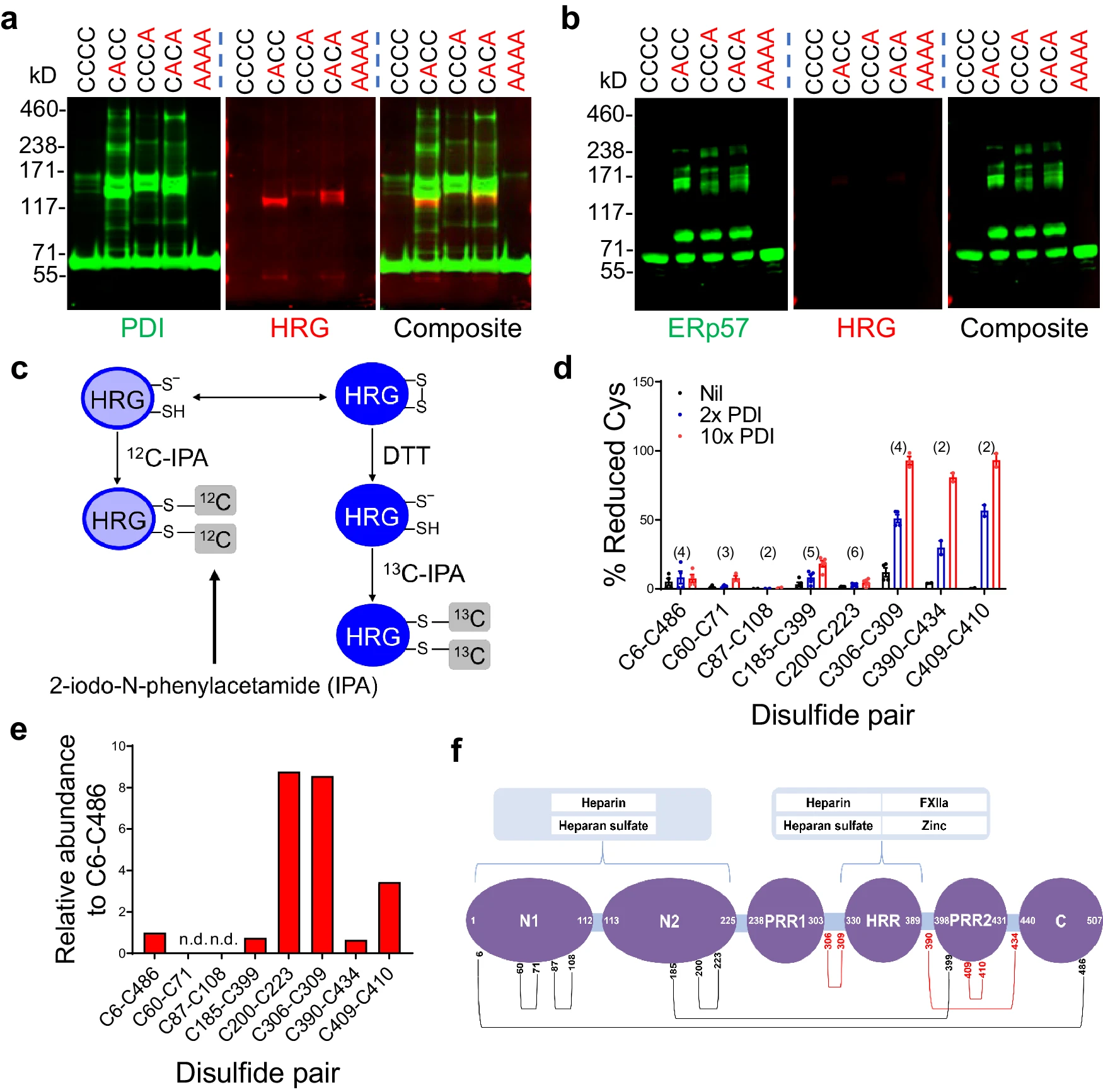
Lv K. et al. Nature Communications. 2024.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Protein Disulfide Bonds Identification and Quantitative Analysis Service
How to order?







