Differences Between Peripheral and Integral Membrane Proteins and Methods for Their Identification
-
TMHMM / Phobius / TOPCONS: Predict transmembrane helices and membrane topology
-
SignalP: Distinguish signal peptides from transmembrane regions
-
UniProt database annotation: Verify whether proteins are annotated as "transmembrane"
-
Detergents: SDS, Triton X-100, NP-40, etc.
-
Organic solvent systems: e.g., methanol/chloroform extraction
-
Membrane protein-specific phase separation kits: e.g., Mem-PER™, CelLytic™, suitable for high-throughput analysis
-
Protein Solubilization: Use appropriate detergents to maintain solubility of membrane proteins
-
Protein Digestion: Performed enzymatically (e.g., trypsin) or via Filter-Aided Sample Preparation (FASP)
-
Enrichment: Target proteins can be enriched through phase separation or immuno-enrichment
-
GO annotation keywords such as "membrane" and "integral component of membrane"
-
Number of predicted transmembrane domains
-
Classification and statistical analysis using UniProt and PANTHER databases
-
All integral membrane proteins are membrane proteins, but not all membrane proteins are integral.
-
Different extraction, enrichment, and mass spectrometry strategies are required for the two types.
-
Preliminary classification can be performed using bioinformatics prediction.
-
Experimentally, optimize lysis buffers and select suitable detergents.
-
In data analysis, integrate domain information and database annotations for accurate classification.
Membrane proteins and integral membrane proteins are critical components of the cellular membrane system, playing essential roles in signal transduction, molecular transport, and cell recognition. Although both types of proteins are associated with biological membranes, they differ significantly in structural characteristics, membrane association mechanisms, and functional properties, resulting in distinct experimental identification strategies.
Differences Between Membrane Proteins and Integral Membrane Proteins
1. Definition and Classification
Membrane proteins include all proteins associated with the structure and function of the cell membrane. Based on their mode of association with the membrane, they are classified into two major categories:
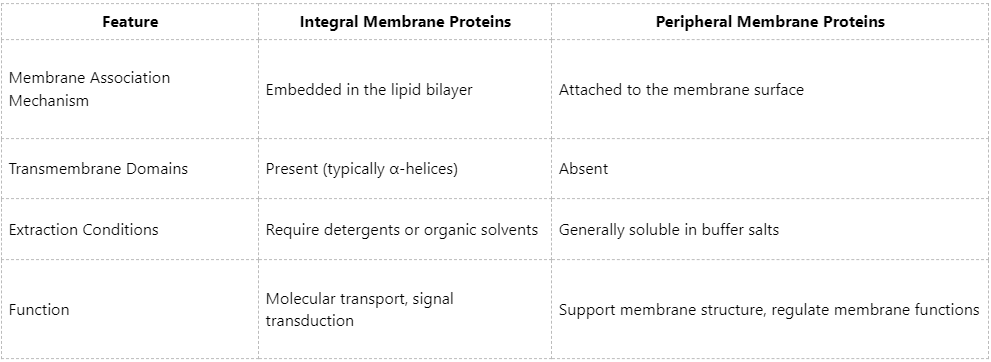
(1) Integral Membrane Proteins
Also referred to as intrinsic membrane proteins, these proteins are embedded within the lipid bilayer and typically contain one or more transmembrane domains, which are primarily hydrophobic α-helices spanning the bilayer.
(2) Peripheral Membrane Proteins
These proteins do not directly insert into the membrane but attach to the inner or outer surface via non-covalent interactions, such as electrostatic interactions, hydrophobic interactions, or protein–protein interactions.
Thus, integral membrane proteins represent a subclass of membrane proteins.
2. Structural Features
Methods for Identifying Membrane Proteins and Integral Membrane Proteins
Due to their high hydrophobicity and low solubility, membrane proteins are among the most challenging targets in proteomics. Identification strategies should be tailored according to the biochemical properties of different protein types.
1. Bioinformatics Prediction (In Silico Analysis)
Before experiments, protein sequences can be analyzed using the following tools to predict transmembrane domains and preliminarily classify proteins as integral membrane proteins:
These tools provide reliable preliminary predictions for protein functional classification and are crucial during proteomics data annotation.
2. Membrane Protein Extraction and Enrichment
(1) Extraction of Integral Membrane Proteins
Because they are embedded in the membrane, disruption of the lipid bilayer is necessary, which can be achieved using:
At MtoZ Biolabs, we combine a modified strong detergent buffer system with mild lysis conditions to enhance extraction efficiency of integral membrane proteins, particularly for low-abundance transmembrane receptors and signaling molecules.
(2) Extraction of Peripheral Membrane Proteins
Mild lysis methods, such as buffer salts or sonication, are commonly used to avoid disrupting the lipid bilayer, allowing peripheral proteins to be released and enriched in the soluble fraction.
3. Identification Techniques: Mass Spectrometry and Membrane Proteomics
Membrane proteomics relies on the high sensitivity and resolution of mass spectrometry, with strict requirements for sample preparation.
(1) Key Steps in Sample Preparation
(2) Common Mass Spectrometry Strategies
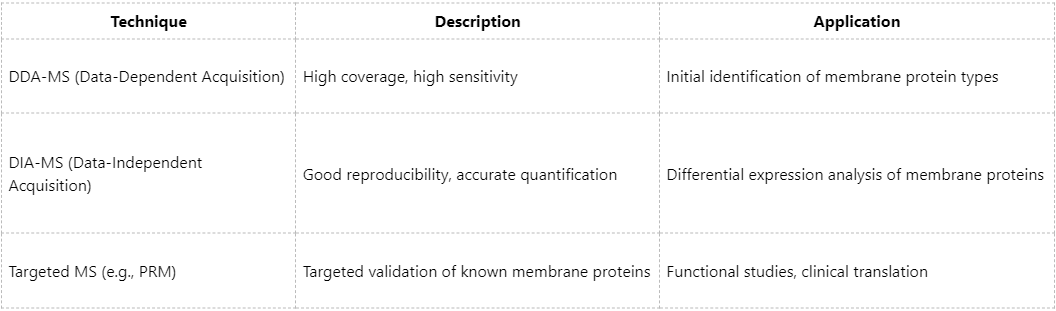
At MtoZ Biolabs, ultra-high-resolution Orbitrap mass spectrometry combined with optimized membrane protein enrichment workflows significantly improves identification throughput and quantitative accuracy, particularly for GPCR and RTK membrane receptor studies.
4. Post-Experimental Data Analysis
Mass spectrometry results can be further analyzed using:
Summary and Prospects
Membrane proteins, especially integral membrane proteins, are central to disease biomarker discovery, drug target screening, and signal pathway analysis. However, their low expression, hydrophobicity, and structural complexity pose significant challenges for proteomics research.
1. Key Distinctions
2. Research Recommendations
For researchers working on membrane proteins or facing technical challenges in extracting and identifying integral membrane proteins, technical support is available from MtoZ Biolabs. We provide comprehensive solutions covering sample preparation, membrane protein enrichment, mass spectrometry analysis, and bioinformatics, facilitating advanced membrane proteomics research.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







