DIA vs. DDA in Label-Free Quantitative Proteomics: A Comparative Analysis
-
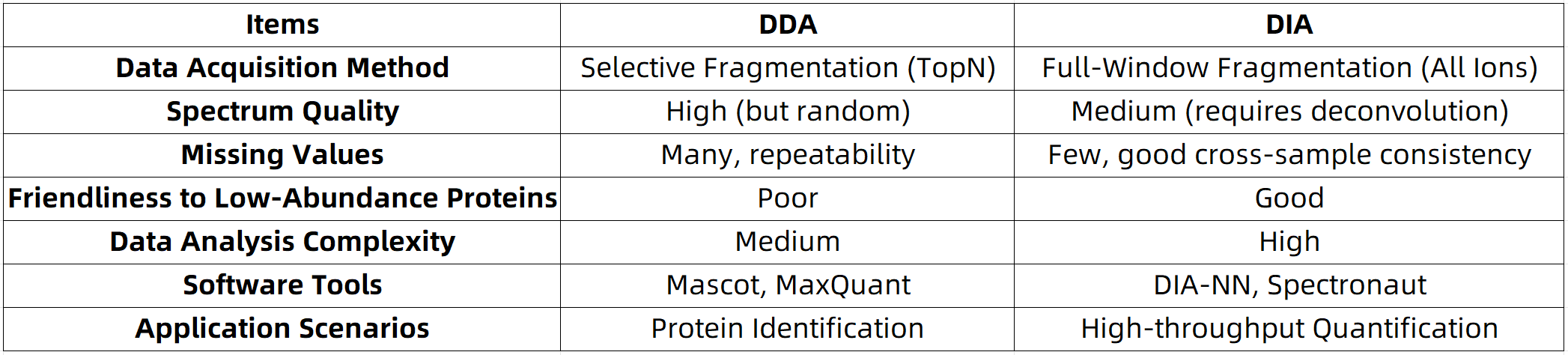
High-quality spectra: By focusing on the most intense signals, DDA ensures a high signal-to-noise ratio, facilitating accurate and confident peptide identification.
-
Efficient database searching: The generated MS/MS spectra are compatible with established database search algorithms such as SEQUEST and Mascot, enabling efficient protein identification workflows.
-
Limited reproducibility: Precursor ions selected for fragmentation can vary across different samples or experimental runs, often leading to missing data points.
-
Reduced sensitivity to low-abundance proteins: High-abundance proteins are preferentially selected, which may result in the underrepresentation or omission of low-abundance species.
-
Constrained dynamic range: In complex biological samples, dominant high-abundance proteins may suppress the detection of less abundant peptides, limiting overall proteomic coverage.
-
High reproducibility and data completeness: Ion selection remains consistent across samples, resulting in significantly fewer missing values.
-
Enhanced detection of low-abundance proteins: The comprehensive acquisition approach reduces bias toward high-abundance precursors.
-
Facilitates the generation of robust and reproducible proteomic spectral libraries.
-
High spectral complexity: The substantial overlap in MS2 spectra necessitates advanced computational tools for accurate deconvolution.
-
Strong reliance on algorithmic processing: Effective data interpretation depends on specialized DIA analysis software, such as Spectronaut, DIA-NN, and EncyclopeDIA.
Label-free quantitative proteomics is widely used to investigate dynamic changes in protein expression within biological systems and plays a critical role in areas such as disease mechanism elucidation, drug target discovery, and biomarker identification. Among label-free quantification strategies, Data-Dependent Acquisition (DDA) and Data-Independent Acquisition (DIA) are the two most commonly employed mass spectrometry acquisition approaches, each characterized by distinct technical features and suited to different applications. This article provides a systematic comparison of these two strategies to aid researchers in selecting the most appropriate method for experimental design.
What Is DDA (Data-Dependent Acquisition)?
1. Brief Principle
DDA operates by prioritizing high-intensity signals during data acquisition. In each MS cycle, the mass spectrometer first conducts an MS1 full scan, then selects the top N most intense precursor ions for MS2 fragmentation based on their signal strength. This strategy yields high-quality MS/MS spectra and is well-suited for protein identification tasks.
2. Advantages
3. Limitations
What Is DIA (Data-Independent Acquisition)?
1. Brief Principle
DIA employs a data-independent acquisition strategy, in which all precursor ions within predefined m/z windows are simultaneously and systematically fragmented, regardless of their intensity. Representative techniques include SWATH-MS and diaPASEF. Unlike DDA, DIA acquires comprehensive and consistent MS2 spectra across the entire mass range, making it well-suited for label-free quantitative proteomic analysis.
2. Advantages
3. Limitations
Comparative Analysis of Core Performance Metrics: DIA vs. DDA
Recommendations for Application Scenario Selection
1. Exploratory Research / Novel Species Investigation
For research aimed at discovering unknown proteins or constructing comprehensive protein atlases, DDA is generally preferred owing to its superior spectral quality and effective database search capabilities.
2. High-Throughput Quantification / Clinical Cohort Analysis
In large-scale cohort analyses and disease biomarker screening—typical high-throughput quantitative applications—DIA is more suitable due to its high data consistency and reduced rates of missing values, facilitating more reliable differential protein identification and statistical evaluation.
3. Mass Spectral Library Construction Strategy
A widely adopted approach involves initially using DDA to generate a high-quality spectral library, followed by DIA for quantification. This “DDA+DIA integrated strategy” combines the strengths of data quality and analytical throughput, and has emerged as one of the prevailing methodologies.
Both DDA and DIA offer distinct advantages. Researchers should make informed decisions based on specific experimental goals, sample characteristics, and available budget. As a next-generation mainstream technique, DIA is gaining increasing prominence in label-free quantitative proteomics. Nevertheless, DDA remains indispensable for spectral library generation and the identification of novel proteins. MtoZ Biolabs is dedicated to delivering high-quality label-free quantitative proteomics services, supporting a broad spectrum of applications including basic research, disease mechanism elucidation, biomarker discovery, and clinical translation.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







