Cryo-Electron Tomography (Cryo-ET) Analysis Service
- Subcellular Structural Imaging: 3D reconstruction of organelles, nuclear pores, mitochondrial cristae, cytoskeletal networks, etc.
- Viral Infection and Replication Mechanisms: Analysis of virus–host interfaces, membrane fusion events, and assembly intermediates.
- Bacterial and Fungal Ultrastructure: Visualization of flagella, secretion systems, and cell wall nanostructures.
- Membrane Protein Complex Localization and Conformational Analysis: Structural insights into transmembrane assemblies, signaling complexes, and channel proteins.
- Nanostructures and Biomaterials: Characterization of synthetic systems, DNA nanostructures, and artificial vesicle systems.
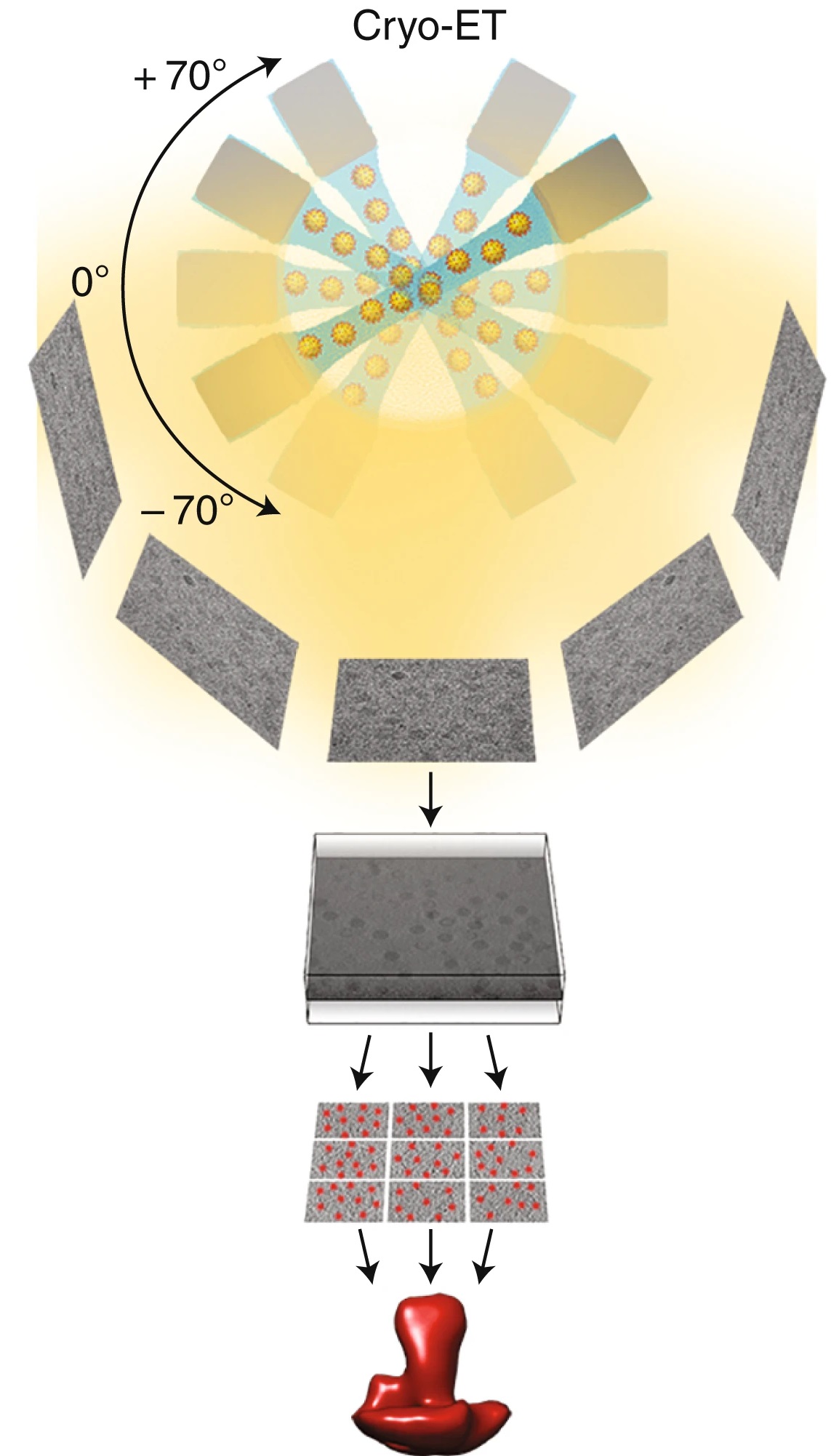
Cryo-Electron Tomography (Cryo-ET) Analysis Service captures high-resolution 3D density maps by imaging vitrified specimens at multiple tilt angles, followed by tomographic reconstruction. This enables in situ structural analysis, subunit localization, conformational classification, and functional annotation of target regions.
In life sciences and structural biology, understanding the 3D conformation of intracellular protein complexes, subcellular structures, and viral particles in a near-native state is essential for elucidating their functional mechanisms. However, traditional structural techniques—such as X-ray crystallography and single-particle cryo-electron microscopy—rely on highly purified or repetitive samples and are limited in capturing spatial information of non-repetitive structures in their native cellular context.
Luque D. Castón JR. Nat Chem Biol. 2020.
Cryo-ET, as an advanced cryo-electron microscopy technique, enables nanoscale three-dimensional reconstruction of vitrified specimens without the need for staining or chemical fixation, preserving the native spatial distribution and conformational states of biomolecules. It is a powerful tool for studying complex systems such as organelle architecture, membrane protein complexes, viral infection pathways, and bacterial nanostructures.
MtoZ Biolabs offers Cryo-Electron Tomography (Cryo-ET) Analysis Service to provide nanoscale 3D reconstruction and analysis of cells, viruses, macromolecular assemblies, and ultrastructures under near-physiological conditions. We deliver end-to-end solutions—from sample preparation and image acquisition to 3D reconstruction and structural visualization—empowering researchers to uncover the in situ structural basis of biological processes.
Technical Principles
Cryo-Electron Tomography (Cryo-ET) is a three-dimensional reconstruction technique that combines cryo-electron microscopy (Cryo-EM) with electron tomography. Its core principles include:
1. Vitrification of Samples: Biological specimens are rapidly frozen into vitreous ice without the use of stains or fixatives, preserving their native ultrastructure as closely as possible.
2. Tilt Series Imaging: Using a transmission electron microscope (TEM), the sample is incrementally tilted, and a series of 2D projection images are acquired at each angle.
3. 3D Tomographic Reconstruction: Using computational methods such as weighted back-projection (WBP) or the simultaneous iterative reconstruction technique (SIRT), the tilt series is reconstructed into a 3D density map (tomogram), revealing the near-native conformation of the target structure.
Analysis Workflow
Cryo-Electron Tomography (Cryo-ET) Analysis Service provided by MtoZ Biolabs typically includes the following steps:
1. Sample Preparation and Vitrification
Samples are plunge-frozen in liquid nitrogen to form vitreous ice and thinned as needed to ensure electron beam penetration.
2. Tilt Series Acquisition
Automated collection of multi-angle images within a ±60° tilt range is performed on a cryo-TEM under low-dose conditions, ensuring high contrast and minimal radiation damage.
3. 3D Reconstruction
Image alignment and reconstruction algorithms are used to generate high-fidelity 3D tomograms from the tilt series.
4. Image Analysis
Segmentation, denoising, structure recognition, and sub-tomogram averaging are applied to enhance resolution and analyze conformational states.
Service Advantages
1. In Situ Imaging: Direct visualization of cells and macromolecules in 3D under near-physiological conditions, preserving native conformations.
2. No Crystallization Required: Ideal for non-crystalline targets such as membrane proteins, viruses, and subcellular structures.
3. High Spatial Resolution: Achieves nanoscale resolution for 3D reconstructions; sub-tomogram averaging can further improve resolution to sub-nanometer levels.
4. Suitable for Heterogeneous Samples: Capable of resolving structurally diverse or conformationally dynamic macromolecular assemblies that lack symmetry or repetition.
Applications
Cryo-ET Analysis Service is ideal for studying heterogeneous, non-purified, or in situ biological systems, with broad applications such as:
FAQ
Q. How is Cryo-ET Different from Conventional Single-Particle Cryo-EM (SPA)?
Cryo-ET does not rely on particle homogeneity or symmetry, making it suitable for asymmetric, heterogeneous, or unpurifiable structures such as organelles, viral entry machinery, or membrane protein networks. SPA, in contrast, is optimized for high-purity, repetitive particles and aims to generate a high-resolution average structure.
Q. What Types of Samples are Suitable for Cryo-ET?
Cryo-ET is compatible with thin cell layers, FIB-milled lamellae, virus particles, bacteria, organelles from eukaryotic cells, membrane protein complexes, nuclear pores, mitochondria, vesicle systems, and other in situ biological targets.
Q. Does the Sample Need to be Purified in Advance?
No. One of Cryo-ET's major strengths is its ability to perform in situ imaging in complex environments. It can directly analyze unpurified cells, tissues, or biological particles.
Case Study
This study used cryo-focused ion beam (cryo-FIB) and cryo-electron tomography (Cryo-ET) technology to resolve the three-dimensional structure of natural chromatin fibers in human T lymphoblasts under in situ conditions for the first time. The study found that these chromatin fibers are not the uniform 30-nanometer one- or two-strand helical structures traditionally believed, but are organized in a loose, variable, zigzag manner by nucleosomes connected by straight chains of DNA.
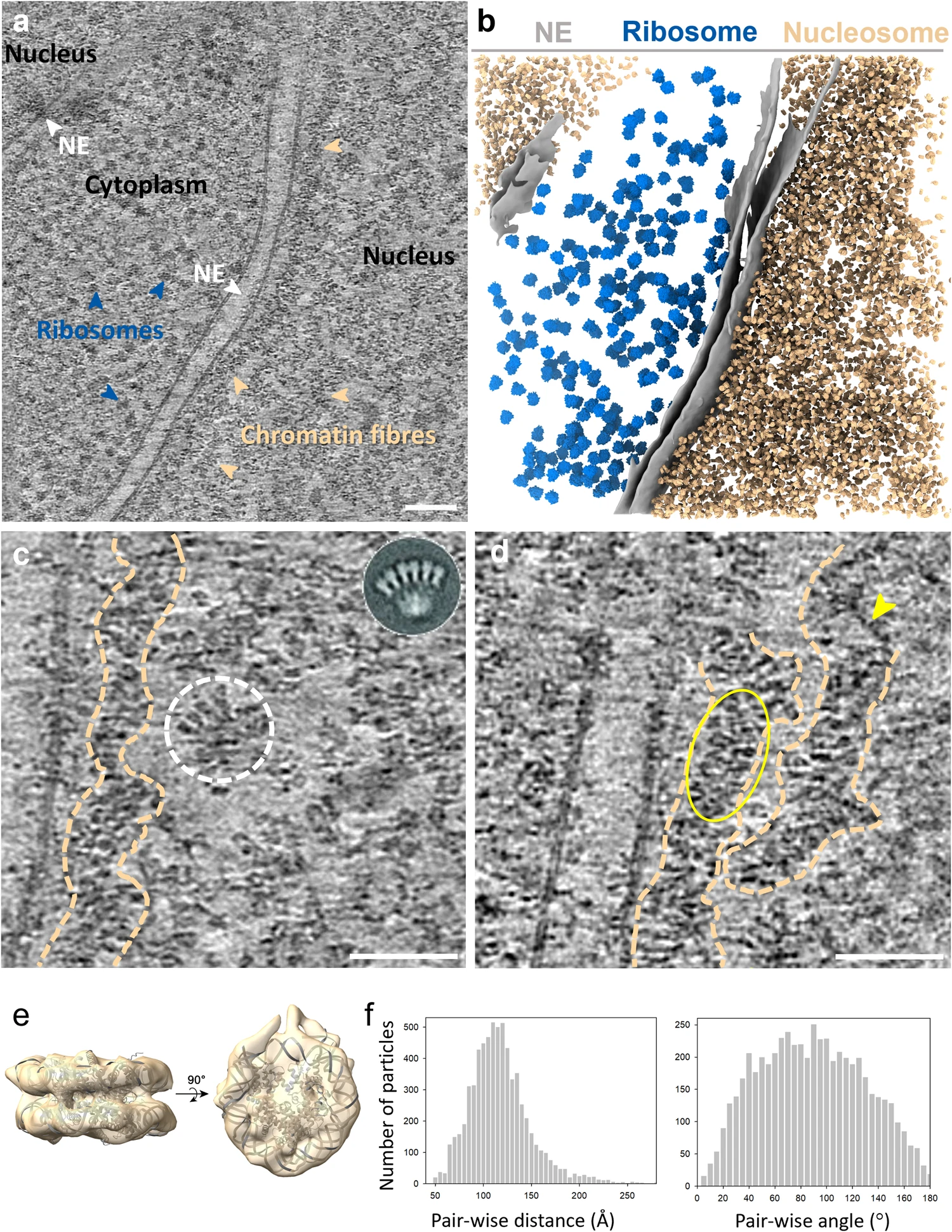
Hou Z. et al. Nat Commun. 2023.
How to order?







