Crosslinking Mass Spectrometry Service
Crosslinking Mass Spectrometry (XL-MS) is a cutting-edge technique that integrates chemical crosslinking with mass spectrometry to study protein structures and interactions at the molecular level. Crosslinking mass spectrometry method stabilizes transient or dynamic protein-protein interactions by creating covalent bonds between amino acid residues in close proximity, preserving their spatial relationships. As a result, XL-MS provides critical insights into the organization, dynamics, and functional mechanisms of protein complexes, making it a transformative tool in structural biology and proteomics.
The principle of crosslinking mass spectrometry involves using chemical crosslinkers to covalently bond specific amino acid residues in close proximity. These crosslinked peptides are then analyzed using high-resolution mass spectrometry, which identifies the linked residues and determines their spatial relationship within the protein or complex. This data provides distance restraints that can be used for structural modeling, revealing conformational changes, interaction surfaces, and the architecture of multi-protein assemblies. XL-MS also excels in capturing both stable and transient interactions, making it highly valuable for studying dynamic biological processes.
Crosslinking mass spectrometry is widely used to define the architecture of large protein complexes, map protein-protein interaction networks in vitro and in vivo, and validate computational or cryo-EM-derived models. Additionally, crosslinking mass spectrometry facilitates the study of conformational dynamics, revealing how proteins fold, assemble, or respond to external stimuli. Its ability to capture interactions in their native or near-native state makes it indispensable for advancing drug discovery, structural biology, and systems biology.
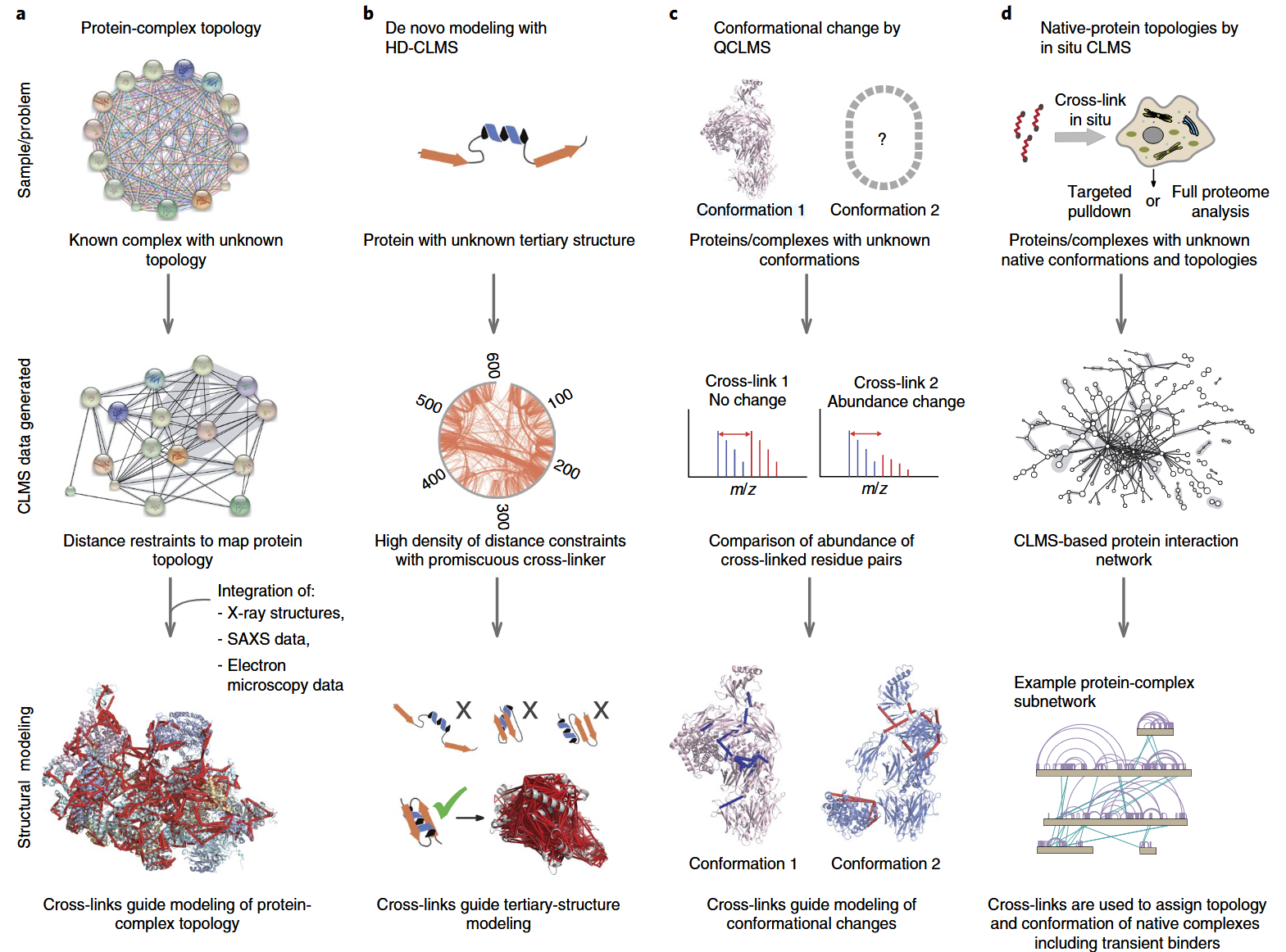
Figure 1. Applications of Crosslinking Mass Spectrometry
Service at MtoZ Biolabs
MtoZ Biolabs offers an advanced Crosslinking Mass Spectrometry Service tailored to uncover protein-protein interaction networks and define the architecture of protein complexes. Utilizing cutting-edge mass spectrometry platforms, high-performance cross-linking reagents, and powerful bioinformatics tools, we provide an end-to-end solution for XL-MS studies.
Our approach covers every critical stage of the crosslinking mass spectrometry workflow, from selecting the most suitable cross-linkers to optimizing sample preparation protocols that preserve native protein interactions. High-resolution mass spectrometry is employed to ensure accurate identification and quantification of crosslinked peptides. Following data acquisition, our bioinformatics experts process the complex datasets to pinpoint crosslinked residues, enabling detailed mapping of interaction sites and structural modeling. With expertise spanning structural characterization and dynamic interaction analysis, our Crosslinking Mass Spectrometry Service delivers comprehensive insights, empowering researchers to unravel the complexities of protein interactions and dynamics in health and disease.
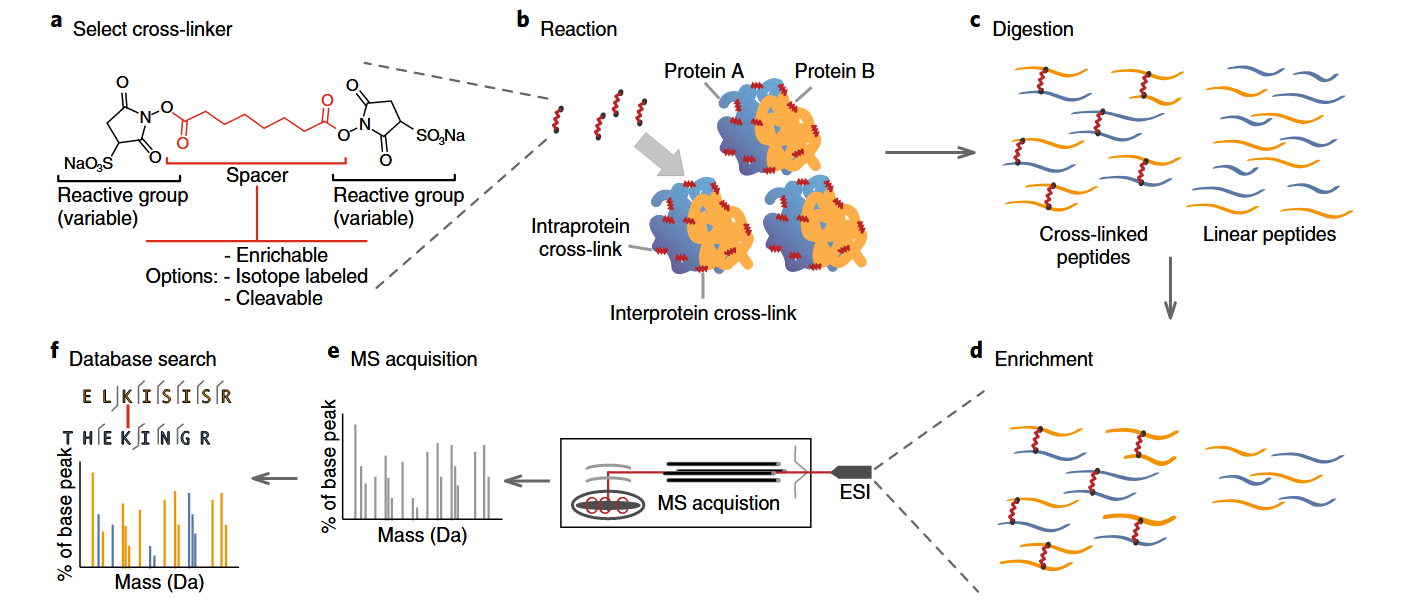
Figure 2. General Workflow of Crosslinking Mass Spectrometry Service
Service Advantages
1. Advanced Analysis Platform
MtoZ Biolabs established an advanced Crosslinking Mass Spectrometry Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. Low Sample Requirement
XL-MS requires only a small amount of protein for analysis, enabling the study of proteins that are difficult to isolate or available in limited quantities.
3. Preservation of Native Protein States
Crosslinking mass spectrometry facilitates the study of protein structures and interactions under near-native conditions, enabling detailed and accurate analysis of protein conformations and interactions with minimal perturbation to their native states.
4. One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
5. High-Data-Quality
Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all crosslinking mass spectrometry data, providing clients with a comprehensive data report.
6. Rapid Turnaround Time
Unlike traditional high-resolution techniques such as cryo-EM or X-ray crystallography, XL-MS experiments can be completed within a few days, making it an efficient option for structural analysis.
Applications
The Crosslinking Mass Spectrometry Service can be used in:
1. Structural characterization of protein complexes.
2. Mapping protein-protein interaction (PPI) networks.
3. Validating computational and cryo-EM models.
4. Studying transient protein interactions.
5. Investigating protein conformational dynamics.
6. Supporting drug target discovery and validation.
Sample Results
Our Crosslinking Mass Spectrometry Service delivers high-quality data and comprehensive analyses, providing clients with clear and actionable insights into protein structures and interactions. Below are examples of the types of results you can expect:
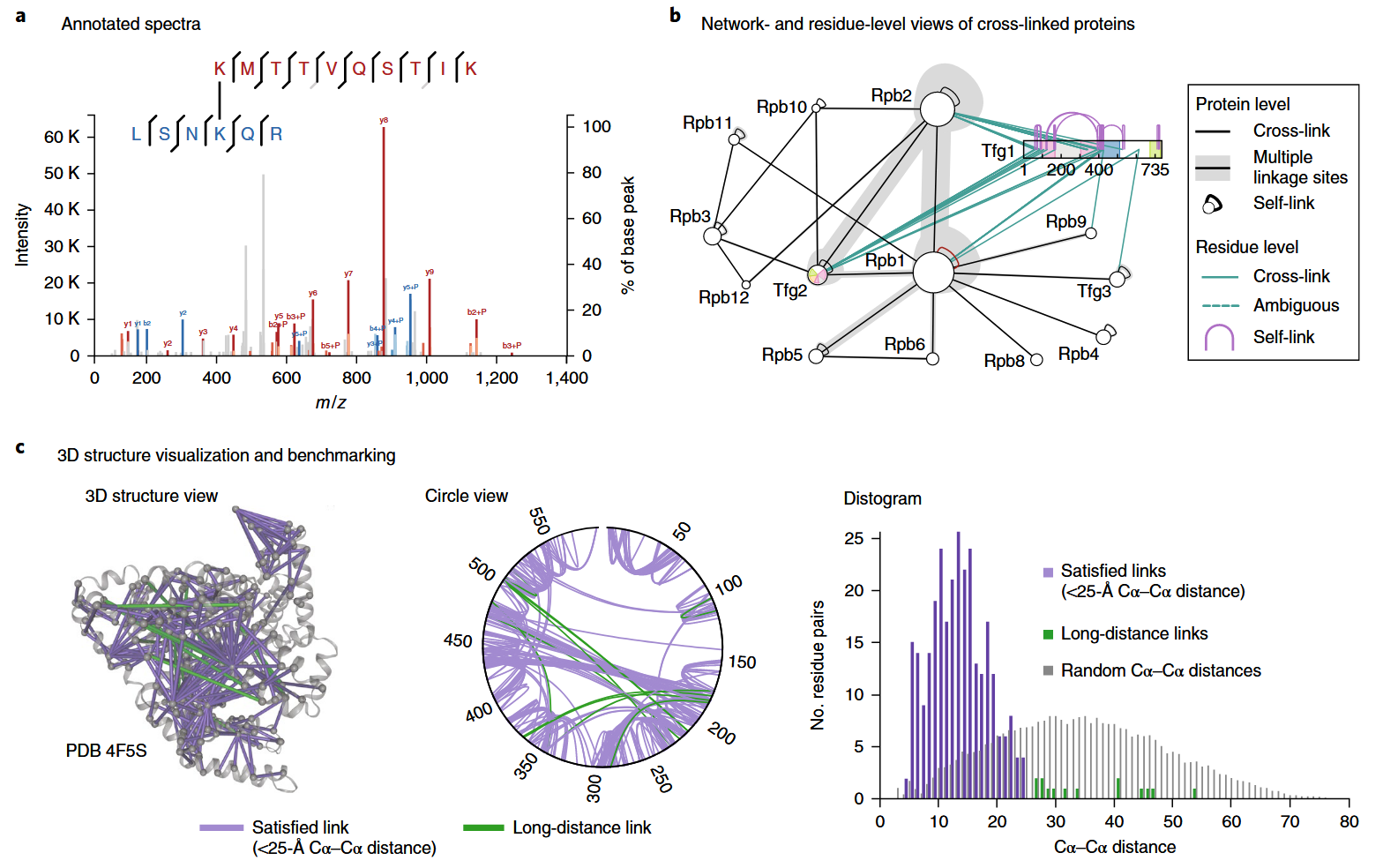
Figure 3. Sample Results of Crosslinking Mass Spectrometry Service
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Crosslinking Protein Interaction Analysis Service
Chemical Crosslinking-Mass Spectrometry (CXMS) Analysis Service
How to order?







