Chemical Crosslinking-Mass Spectrometry (CXMS) Analysis Service
Chemical Crosslinking-Mass Spectrometry (CXMS) Analysis Service is a specialized service that integrates chemical crosslinking reactions with high-resolution mass spectrometry to study protein structure and interactions. It is designed to elucidate spatial conformations of proteins or protein complexes and map their interaction interfaces.
Understanding protein–protein interactions and their three-dimensional structures is critical for uncovering cellular functional mechanisms, signaling pathway regulation, and the modes of action of disease-related protein complexes. However, conventional structural biology techniques—such as X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy—often face limitations when dealing with dynamic, heterogeneous, or low-abundance protein assemblies. As a structural biology strategy that combines covalent crosslinking with high-resolution mass spectrometry, Chemical Crosslinking-Mass Spectrometry (CXMS) offers high sample compatibility, throughput, and sensitivity to conformational details. It has been increasingly applied in proteomics, interactomics, and structural-functional studies.
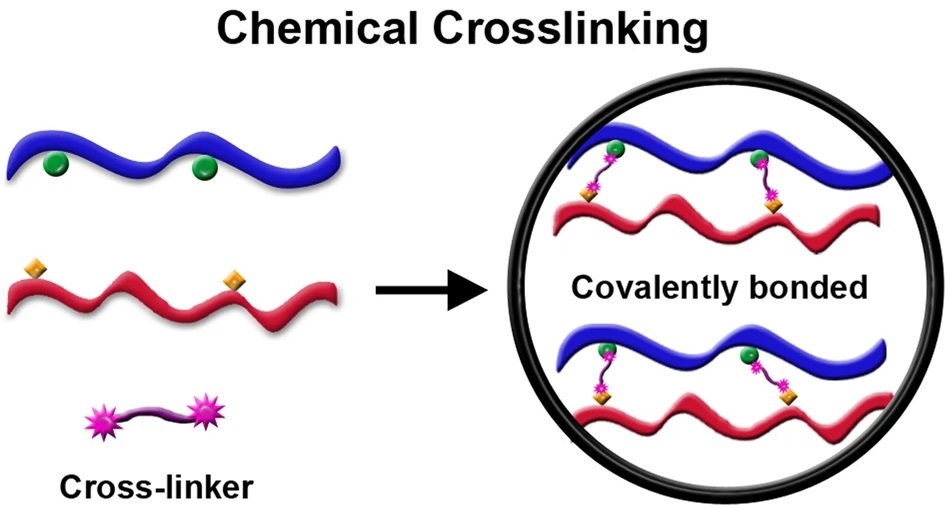
Ramachandran R. et al. NPG Asia Mater. 2019.
The core principle of CXMS lies in covalently linking spatially proximal amino acid residues within or between proteins using controlled crosslinkers, effectively “freezing” the spatial topology of protein complexes at the moment of reaction. Mass spectrometry is then used to identify the crosslinked peptides, allowing researchers to infer interaction sites and spatial constraints either between proteins or within a single protein. This technique is applicable for probing structural contact relationships in protein–protein, protein–DNA/RNA complexes, and multi-subunit protein assemblies.
Leveraging an advanced crosslinking mass spectrometry platform, MtoZ Biolabs offers Chemical Crosslinking-Mass Spectrometry (CXMS) Analysis Service to precisely capture intramolecular and intermolecular spatial constraints. This enables structural characterization and interaction interface mapping of proteins and complexes under near-physiological conditions, helping clients gain deep insights into protein conformational states, dynamic changes, and interaction networks.
Analysis Workflow
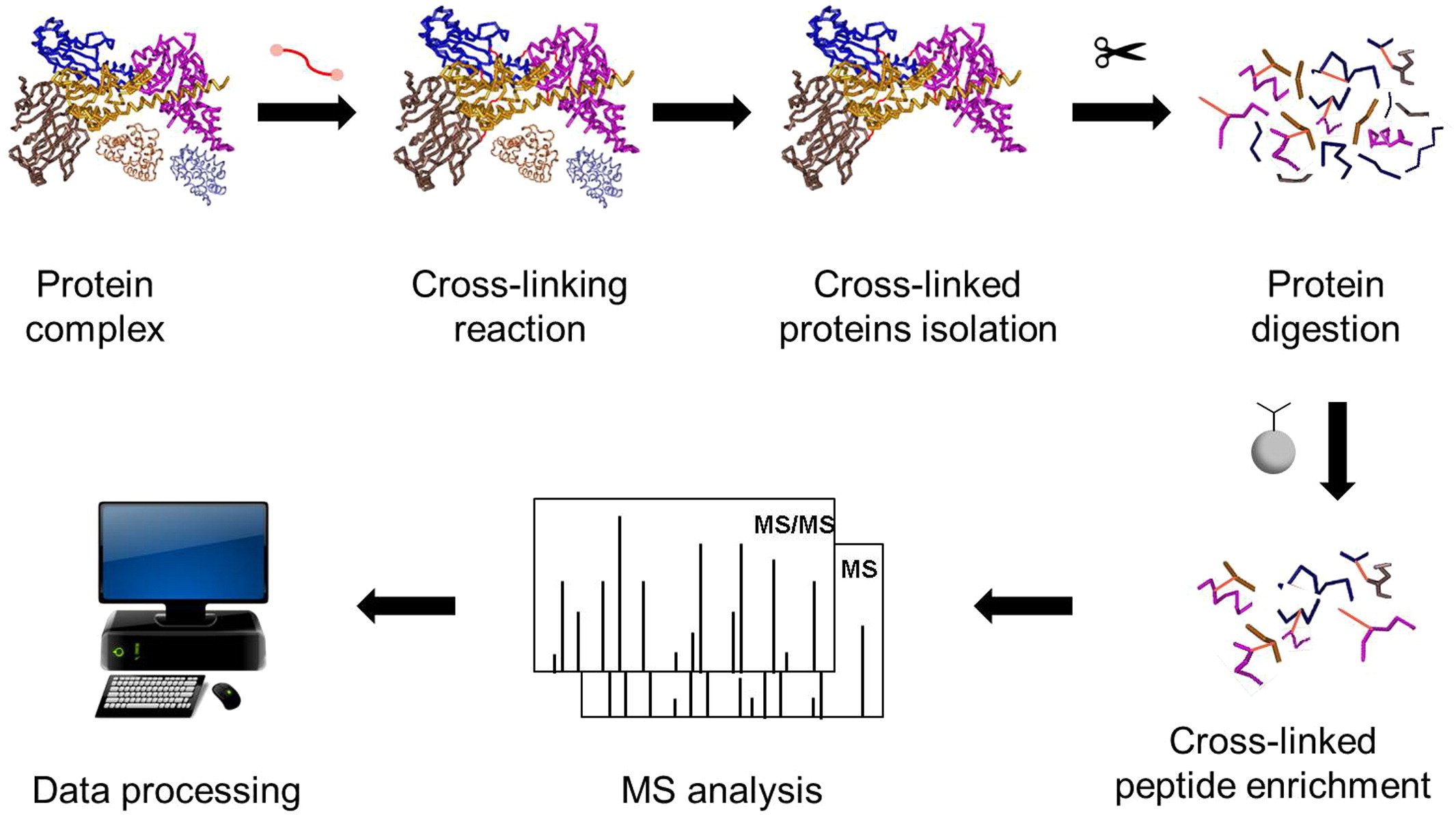
Tran BQ. et al. Biochim Biophys Acta. 2016.
The general workflow of Chemical Crosslinking-Mass Spectrometry (CXMS) Analysis Service is as follows:
1. Crosslinking Reaction
Select appropriate crosslinkers and optimize pH, buffer conditions, and reaction time to perform the chemical crosslinking reaction.
2. Proteolysis and Crosslinked Peptide Enrichment
Use trypsin or a combination of proteases to improve peptide coverage, followed by enrichment of crosslinked peptides.
3. High-Resolution Mass Spectrometry Analysis
Detect crosslinked peptides using a high-resolution mass spectrometry platform.
4. Data Analysis and Spatial Modeling
Employ specialized algorithms to identify crosslinked peptides and analyze crosslinking sites. Integrate crosslinking data into structural modeling for 3D structure construction, conformational filtering, and interaction interface reconstruction.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Chemical Crosslinking-Mass Spectrometry (CXMS) Analysis Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Chemical Crosslinking-Mass Spectrometry (CXMS) Analysis data, providing clients with a comprehensive data report.
4. Broad Sample Compatibility: Suitable for analyzing complexes, membrane proteins, and low-abundance samples that are difficult to resolve using traditional structural techniques.
5. In Situ Capture of Dynamic Conformations: Enables the recording of protein conformational states in solution, revealing structural rearrangements during transitional processes.
Sample Submission Suggestions
Sample Types: Recombinant proteins, native complexes, cell lysates, immunoprecipitation products, etc.
Buffer Requirements: Avoid buffers containing Tris or other amine-containing components whenever possible.
Additional Notes: If you are uncertain about the sample submission requirements, please feel free to contact our technical support team.
Applications
Chemical Crosslinking-Mass Spectrometry (CXMS) Analysis Service can be applied in a variety of research areas, including:
Protein 3D Structure Modeling
Captures the folding patterns of proteins or complexes in their native state and complements missing flexible regions in EM or crystallographic structures.
Protein–Protein Interaction Mapping
Accurately identifies interacting residue pairs, reveals regulatory mechanisms, and supports functional validation and target design.
Conformational Dynamics of Protein Complexes
Compares structural changes under different physiological conditions, providing insights into functional regulation and conformation-dependent mechanisms.
Structural Validation of Drug Targets
Precisely characterizes the binding regions between small molecules and target proteins, offering experimental support for structure-based drug optimization.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Related Services
Protein-Protein Interaction Analysis Service
How to order?







