Comprehensive Untargeted Metabolomics Solutions
Untargeted metabolomics is a comprehensive, unbiased analytical strategy designed to perform full-spectrum analysis of all detectable small molecules in biological samples without prior selection of target metabolites. It combines high-resolution mass spectrometry technologies (such as liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS)) with various chromatographic separation techniques (such as reversed-phase chromatography (RPLC) and hydrophilic interaction chromatography (HILIC)), enabling both qualitative and semi-quantitative analysis of known and unknown metabolites in samples.
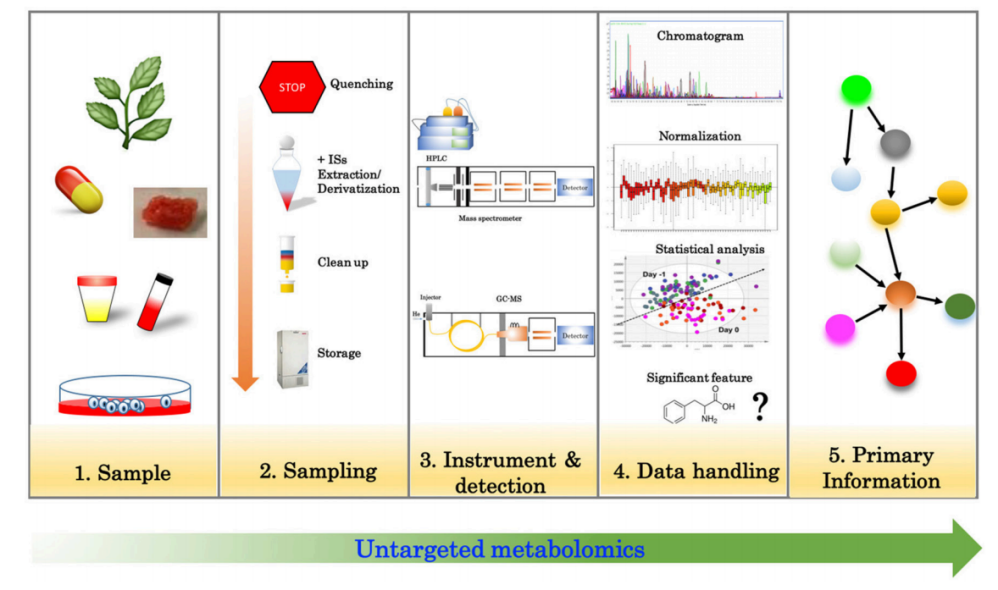
Khoomrung, S. et al. Front Pharmacol. 2017.
Figure 1. Overview of the Steps Involved in Untargeted Metabolomics
During the analysis, untargeted metabolomics covers multiple important metabolite classes such as amino acids, organic acids, lipids, cholesterol, fatty acids, sugars, and nucleotides. Through efficient sample processing and precise instrumentation, this technique allows for comprehensive monitoring of metabolite changes in different physiological or disease states. Untargeted metabolomics is widely used in basic medical research, clinical studies, microbiology, and nutritional science.
Sample Analysis
1. Plasma Metabolomics
Plasma is one of the most commonly used sample types in metabolomics research as it reflects the metabolic state of multiple organs and systems in the body. Plasma contains a wide variety of metabolites and has a large dynamic range, making it commonly used in disease mechanism research, biomarker screening, and drug action monitoring. However, due to the high content of proteins and lipids in blood, the challenge in sample preparation lies in efficiently extracting low-abundance metabolites and removing high-abundance interfering components. To address this, plasma sample processing commonly combines organic solvent protein precipitation with multi-stage purification strategies. For example, methanol, acetonitrile, or their mixtures are used to precipitate plasma proteins, followed by solid-phase extraction (SPE), membrane filtration, or online protein removal systems to enhance metabolite recovery and signal-to-noise ratio. For metabolites with significant polarity differences, a biphasic extraction method (such as the MeOH/MTBE system) is used to separate polar and non-polar metabolites, enabling parallel analysis of lipids and hydrophilic small molecules.
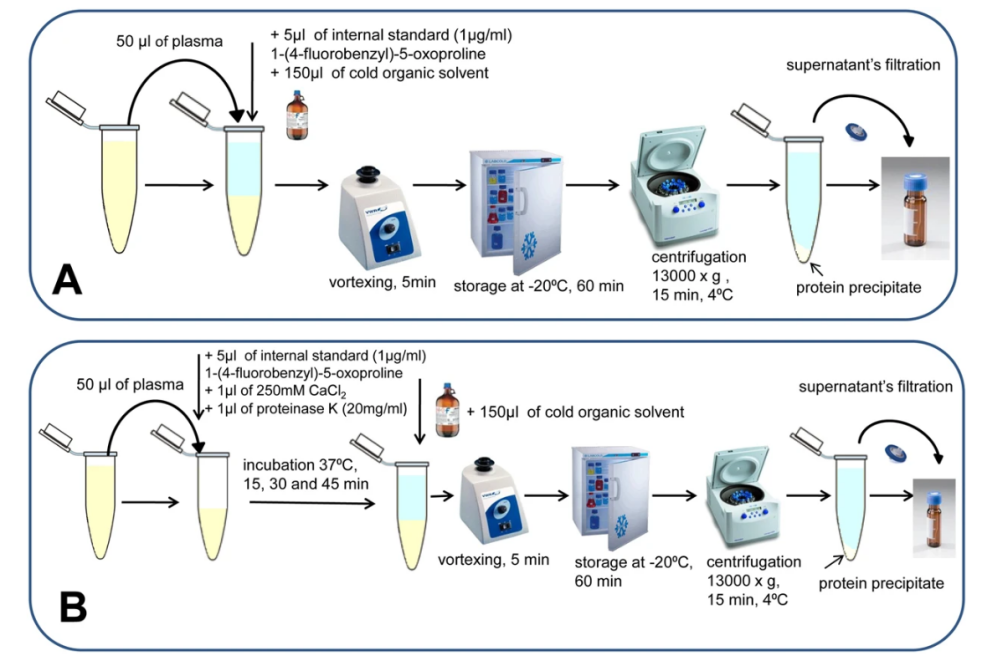
Wawrzyniak, R. et al. Sci Rep. 2018.
Figure 2. Plasma Preparation Procedures in Plasma Metabolomics
In terms of separation and detection, untargeted metabolomics utilizes high-resolution liquid chromatography-mass spectrometry (LC-HRMS) for metabolite analysis. Using multi-mode separation strategies such as reversed-phase chromatography (RPLC) and hydrophilic interaction chromatography (HILIC), it maximizes metabolite coverage to ensure comprehensive detection of low-abundance metabolites. The mass spectrometry component typically employs high-resolution mass spectrometers like Orbitrap or Q-TOF to precisely measure metabolite mass-to-charge ratios (m/z) and perform preliminary structural identification. Combined with data-dependent acquisition (DDA) and data-independent acquisition (DIA) modes, the method enhances the ability to identify unknown metabolites and ensures the completeness and accuracy of the data.
2. Feces Metabolomics
Fecal samples offer unique advantages in untargeted metabolomics, as they reflect both host and microbial metabolic activities, making them highly valuable for studying gut microbiome function, inflammatory states, and metabolic disorders. Processing fecal samples requires ensuring metabolite representativeness while minimizing interference from sample impurities. The first step is usually freezing and grinding or homogenizing to effectively release metabolites. During extraction, polar or mixed organic solvents (such as methanol, water, isopropanol) are used to recover both polar and non-polar metabolites. Depending on the research requirements, single-phase or biphasic extraction strategies can be selected. High-speed centrifugation and filtration remove particulate matter, and some protocols also involve freeze-drying to concentrate metabolites and enhance detection sensitivity.
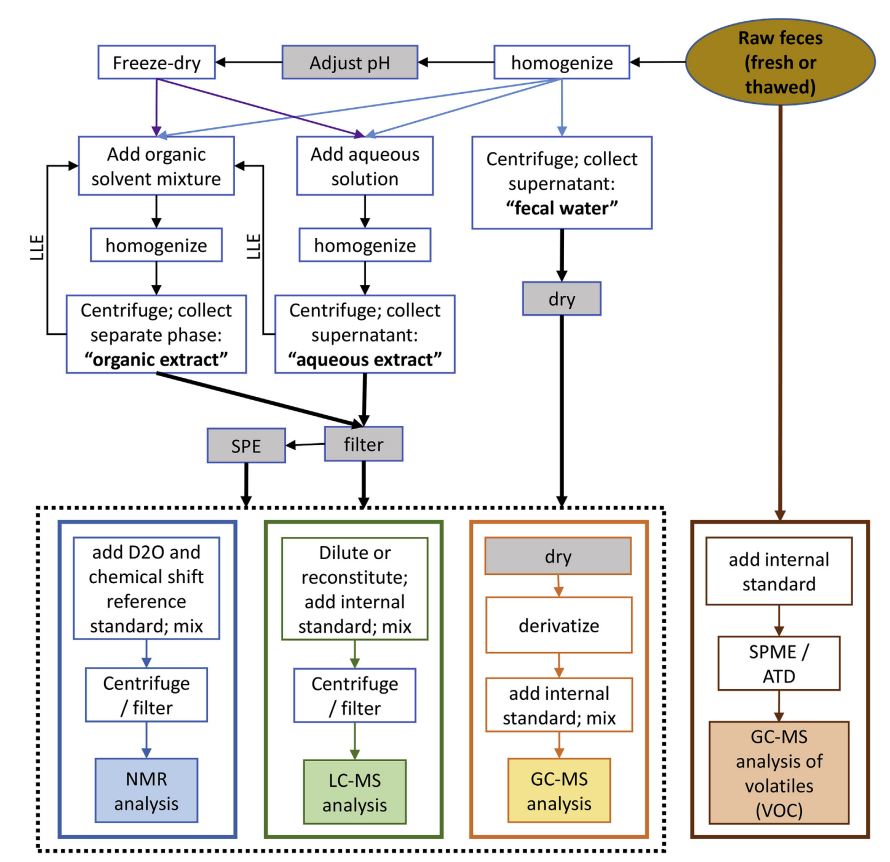
Karu, N. et al. Anal Chim Acta. 2018.
Figure 3. Fecal Sample Preparation Prior to Analysis by Various Platforms
For separation and detection, fecal metabolomics typically combines reversed-phase chromatography (RPLC) and hydrophilic interaction chromatography (HILIC) with high-resolution mass spectrometry (e.g., Orbitrap or Q-TOF) to perform full-scan analysis. This approach covers a broad range of metabolites, including short-chain fatty acids, bile acids, indole compounds, lipids, phenylpropanoids, and microbial-specific metabolites. Due to the complex matrix of fecal samples and large inter-individual variation, data analysis often requires the use of stable internal standards, batch effect correction, and multivariate statistical methods to ensure result comparability and biological interpretation. Fecal metabolomics has broad applications in inflammatory bowel disease, metabolic syndrome, liver-gut axis research, and more, serving as a critical tool for connecting the microbiome with host metabolism.
3. Bacterial Metabolomics
Bacterial metabolomics primarily aims to explore microbial metabolic pathways, strain functions, and the response of microbes to environmental or host stimuli. Bacterial metabolomics is typically divided into intracellular and extracellular metabolite analysis. For intracellular metabolites, rapid termination of metabolism (e.g., using cold solvents) is followed by cell disruption using glass beads or freeze-thaw methods to release metabolites. For extracellular metabolites, they are extracted from bacterial culture supernatants, typically after centrifugation to remove cells, followed by concentration or solid-phase extraction (SPE) purification.
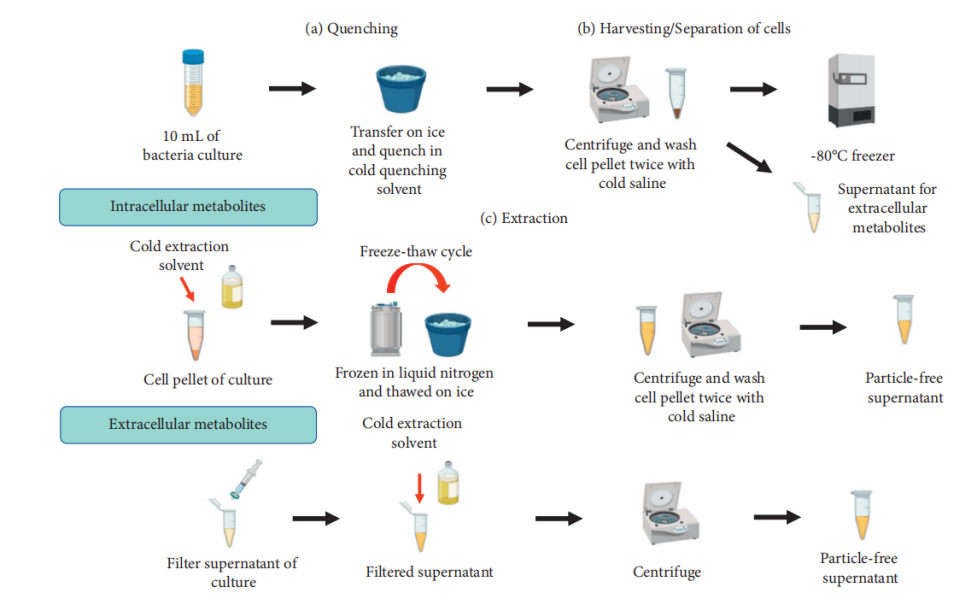
Mohd, Kamal, K. et al. Biochem Res Int. 2022.
Figure 4. Bacterial Sample Preparation for Bacterial Metabolomics
Bacterial samples contain a diverse range of metabolites, including organic acids, short-chain fatty acids, amino acids, alcohols, indole derivatives, and secondary metabolites (such as antibiotics and pigments). These metabolites are separated using HILIC or RPLC chromatographic techniques, followed by high-resolution mass spectrometry (e.g., Orbitrap or Q-TOF) for comprehensive analysis. As bacterial metabolites are often species-specific, data annotation is typically carried out using specialized microbial metabolite databases (e.g., GNPS, KEGG, BioCyc) for comparison and pathway reconstruction. This analysis not only allows for the study of bacterial functions and metabolic pathways but also reveals bacterial responses to external stimuli (e.g., antibiotics), making it a valuable tool for microbiome functional screening, metabolic engineering optimization, and microbiome interaction studies.
Metabolite Analysis
Untargeted metabolomics analysis covers a wide range of metabolites, encompassing various molecular classes. Common analysis components include:
Lipid Analysis: Such as fatty acids , phospholipids, triglyceride , and cholesteryle sters , analyzed using LC-MS or GC-MS for separation and identification. Lipidomics a nalysis involves the comprehensive analysis of lipid, including fatty acids, phospholipids, sphingolipids, triglycerides, cholesterol , and their derivatives. Lipidomics helps to study lipid metabolism, membrane dynamics, and the involvement of lipids in various diseases like cardiovascular disorders, metabolic syndromes, and neurological conditions.
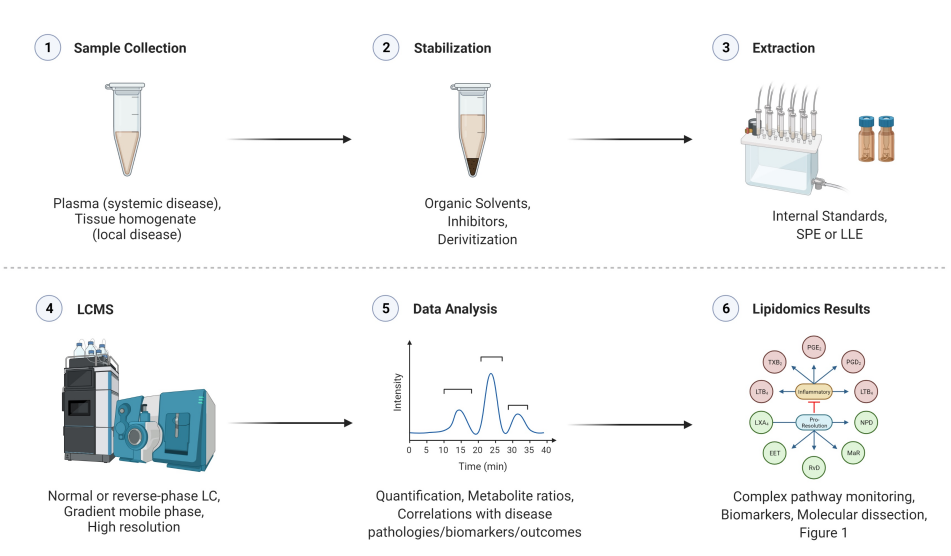
Ahluwalia, K. et al. Metabolites. 2022.
Figure 5. Analysis Workflow of Lipidomics
Amino Acid Analysis: Quantification of 20 common amino acids and their derivatives. Amino acids are the building blocks of proteins and play essential roles in neurotransmission and metabolic regulation.
Bile Acids Analysis: Examining the changes in primary and secondary bile acids, using LC-MS technology for structural identification and quantification. This is widely used in lipid metabolism, liver function, and gut health research.
Metabolomics Pathway Analysis Service
Metabolomics pathway analysis is a critical step in untargeted metabolomics, aimed at mapping detected metabolites to known biological metabolic pathways to identify which pathways are significantly altered under specific conditions. This process typically involves metabolite annotation, pathway matching, enrichment analysis, and impact factor evaluation. By integrating metabolite changes, pathway topology, and network position, it identifies perturbed pathways, assisting in biological mechanism interpretation and providing insights for disease research, drug development, and nutritional interventions.
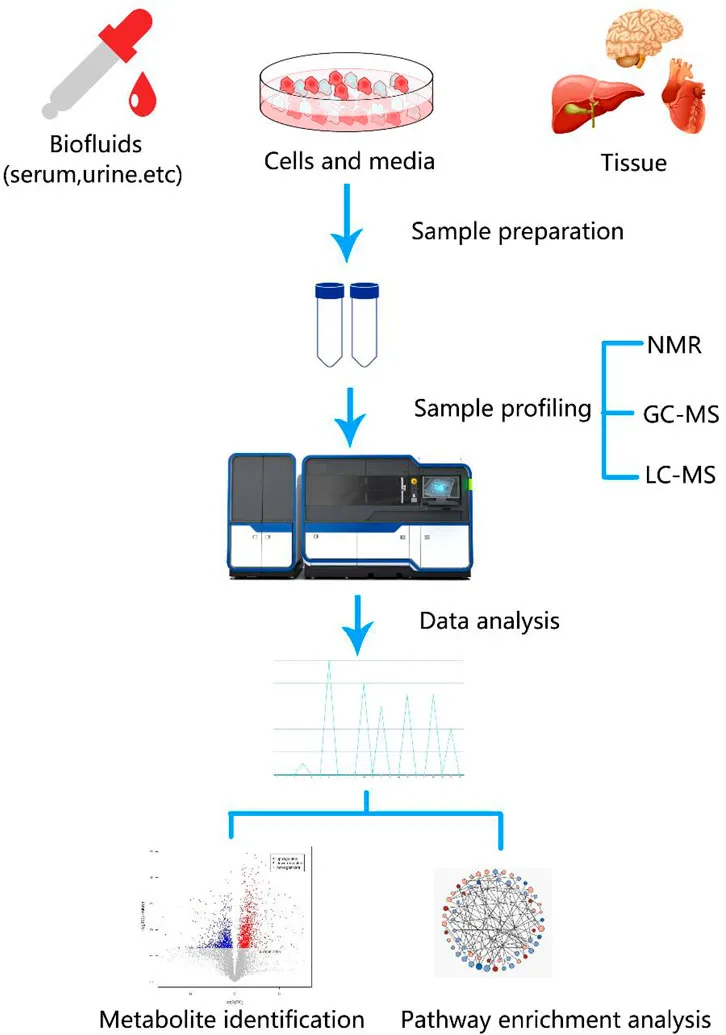
Han, J. et al. Front Mol Biosci. 2021.
Figure 6. Workflow of Metabolomics Pathway Analysis
Advantages
1. Supports multiple sample types for metabolomics analysis, including plasma, feces, bacteria, cells, and tissues.
2. High-resolution mass spectrometry platforms (Orbitrap, Q-TOF) for precise metabolite identification.
3. Multi-mode chromatography techniques (RPLC, HILIC, GC-MS) ensure comprehensive metabolite coverage.
4. Comprehensive database support (KEGG) for metabolite annotation and pathway enrichment.
5. Visualization tools help intuitively interpret metabolite changes and metabolic pathway disturbances.
Limitations
1. Metabolite data are semi-quantitative; absolute quantification requires targeted metabolomics methods.
2. Due to the diversity of metabolites, some novel or unknown metabolites may not be accurately identified.
3. Sample complexity (e.g., feces, bacterial samples) may affect data comparability, requiring optimized processing protocols.
4. Sample matrix differences (e.g., individual variation, environmental factors) may introduce variability, requiring strict control of experimental conditions.
About MtoZ Biolabs
MtoZ Biolabs is a specialized service provider in multi-omics research and high-end mass spectrometry analysis. We are committed to offering high-quality, reproducible, and customizable scientific support to global life science researchers. With advanced mass spectrometry platforms (including Orbitrap Fusion Lumos, Q-Exactive HF, Triple Quad) and a skilled bioinformatics team, we cover the complete workflow from sample processing and metabolite detection to in-depth biological interpretation.
Feel free to contact us for customized metabolomics research solutions, enabling more efficient research and deeper discoveries.
How to order?







