Comprehensive Guide to 4D Label-Free Quantitative Proteomics Workflow
-
Liquid chromatography gradient of 90–120 minutes at nanoflow scale.
-
MS/MS acquisition rate exceeding 100 Hz.
-
Include QC samples to assess MS performance
-
Evaluate peptide length distribution, MS1 signal intensity, and identification saturation
-
Mitigate batch effects via internal standards and normalization techniques
-
Fold change ≥ 1.5 or ≤ 0.67
-
p-value < 0.05 (FDR correction optional)
-
Supported by at least two unique peptides
-
High Sensitivity: Detection limits as low as picogram-level proteins.
-
Comprehensive Bioinformatics Support: End-to-end pipeline covering differential analysis, pathway enrichment, and network modeling.
-
Expert Team: Over a decade of hands-on experience in proteomics R&D and service.
-
Professional Reporting: Delivery of raw data, statistical outputs, high-quality visualizations, and comprehensive interpretation reports.
In the post-genomic era, proteomics has emerged as a pivotal approach for elucidating biological processes, uncovering disease mechanisms, and discovering biomarkers. Specifically, label-free quantitative proteomics (LFQ) has gained widespread application in both basic and translational biomedical research due to its high throughput and the advantage of eliminating the need for costly labeling reagents. With the rapid advancement of mass spectrometry technologies, 4D-based label-free quantification platforms have been developed, offering enhanced quantification accuracy, sensitivity, and proteome coverage, establishing themselves as leading high-end solutions in modern proteomics.
Definition of 4D Label-Free Quantitative Proteomics
4D LFQ integrates four dimensions of separation and identification, namely:
1. Chromatographic retention time (RT)
2. Mass-to-charge ratio (m/z)
3. Ion mobility (IM)
4. Signal intensity
This approach is implemented on the Bruker timsTOF Pro platform utilizing the Parallel Accumulation–Serial Fragmentation (PASEF) acquisition strategy. It enables rapid and sensitive peptide identification and quantification, significantly enhancing proteome depth and data reproducibility.
Detailed Experimental Workflow: From Sample to Data
1. Sample Preparation
(1) Selection of sample type: commonly used sources include cells, tissues, and biological fluids such as serum, plasma, and cerebrospinal fluid.
(2) Protein extraction and quantification: total proteins are efficiently extracted using RIPA or SDT lysis buffers and quantified via the BCA assay.
(3) Enzymatic digestion: proteins are digested using FASP or in-solution methods; trypsin is preferred for its high efficiency and compatibility with PASEF.
(4) Desalting and cleanup: C18 solid-phase extraction is employed to remove contaminants and enhance MS signal quality.
2. Mass Spectrometry Acquisition
(1) Instrumentation: Bruker timsTOF Pro or timsTOF HT platforms equipped with PASEF acquisition mode.
(2) LC-MS parameters:
3. Data Analysis
(1) Database search: data are processed using MaxQuant or Spectronaut and searched against UniProt or SwissProt databases.
(2) Quantification strategy: MS1-level LFQ algorithms quantify changes in protein abundance based on peak area integration.
(3) Statistical analysis: tools such as R and Perseus are used for differential analysis and visualization via volcano plots, heatmaps, etc.
Advantages of 4D Label-Free Quantitative Proteomics
1. Enhanced Resolution via Ion Mobility
Incorporation of ion mobility as a fourth dimension enables resolution of overlapping peptide signals that cannot be distinguished in conventional LC-MS, thereby improving quantification precision.
2. PASEF Enables Ultra-High Scan Rates
By combining parallel ion accumulation with serial fragmentation, scan rates surpass 100 Hz, dramatically increasing protein identification numbers and spectral data density.
3. High Throughput with Minimal Sample Input
Comprehensive proteome coverage can be achieved using as little as 100–200 ng of peptides, making it particularly suitable for precious or limited clinical specimens.
4. Label-Free Strategy Avoids Batch Effects
Samples from different experimental batches can be analyzed independently without chemical labeling (e.g., isotopes, TMT/iTRAQ), reducing cost and enhancing data reproducibility.
Broad Applications Across Research Areas
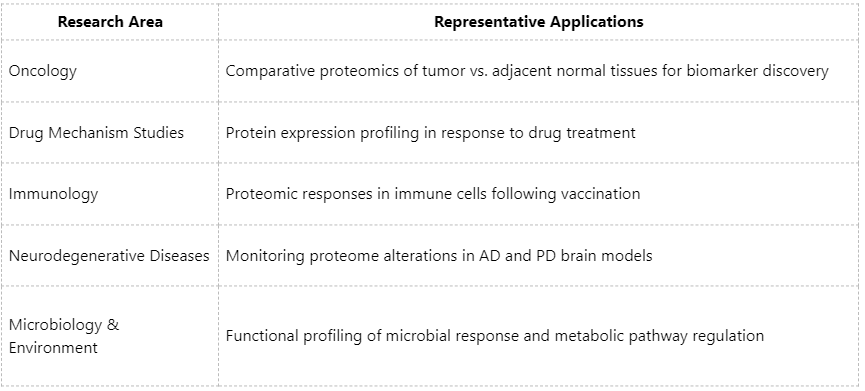
Key Considerations for Data Analysis
1. Quality Control Recommendations
2. Criteria for Identifying Differentially Expressed Proteins
Advantages of MtoZ Biolabs Proteomics Solutions
At MtoZ Biolabs, we integrate the high-resolution timsTOF Pro platform with in-house optimized protocols for protein digestion and purification to offer premium 4D label-free quantitative proteomics services. Key features include:
As mass spectrometry technology continues to evolve and data processing algorithms improve, 4D label-free quantitative proteomics is becoming an indispensable tool in life sciences. Its high throughput, sensitivity, label-free nature, and compatibility with diverse datasets make it highly promising for basic research, mechanistic studies, and drug discovery. For reliable, high-quality 4D proteomics services, we invite you to collaborate with MtoZ Biolabs, we are committed to supporting your research with robust platforms and scientific rigor.
How to order?







